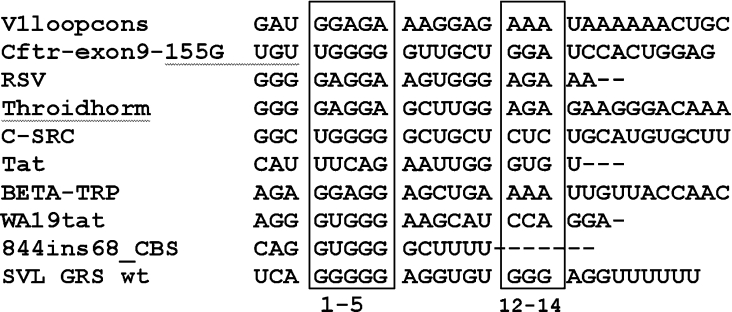
Figure 8. Alignment of hnRNP F- and hnRNP H-binding sites.
Sequences from the indicated genes implicated as binding to hnRNP F or hnRNP H/H2 were taken from the references indicated in the Discussion. The genes are: V1loopcons, consensus for Exon 6D splicing regulatory region of divergent HIV-1 isolates; Cftr-exon9-155G, natural mutation 155G of cystic fibrosis transmembrane regulator Exon 9; RSV, negative regulator of splicing element from rous sarcoma virus; Throidhorm, TRα2 isoform of thyroid hormone receptor gene c-erbAα; C-SRC, intronic splicing enhancer downstream of the N1 5′ splice site of the c-src N1 exon; Tat, a second exonic splicing silencer of tat exon 2 within HIV type I; BETA-TRP, exonic splicing silencer of the rat β-tropomyosin gene; WA19tat, high-affinity hnRNP A1-binding sequence identified by SELEX (systematic evolution of ligands by exponential enrichment); 844 ins68_CBS, 3′ splice site in 884ins68 polymorphism of the cystathionine β-synthase gene. The sequences between nucleotides 1–5 and 12–14 of the GRS are in the boxes.