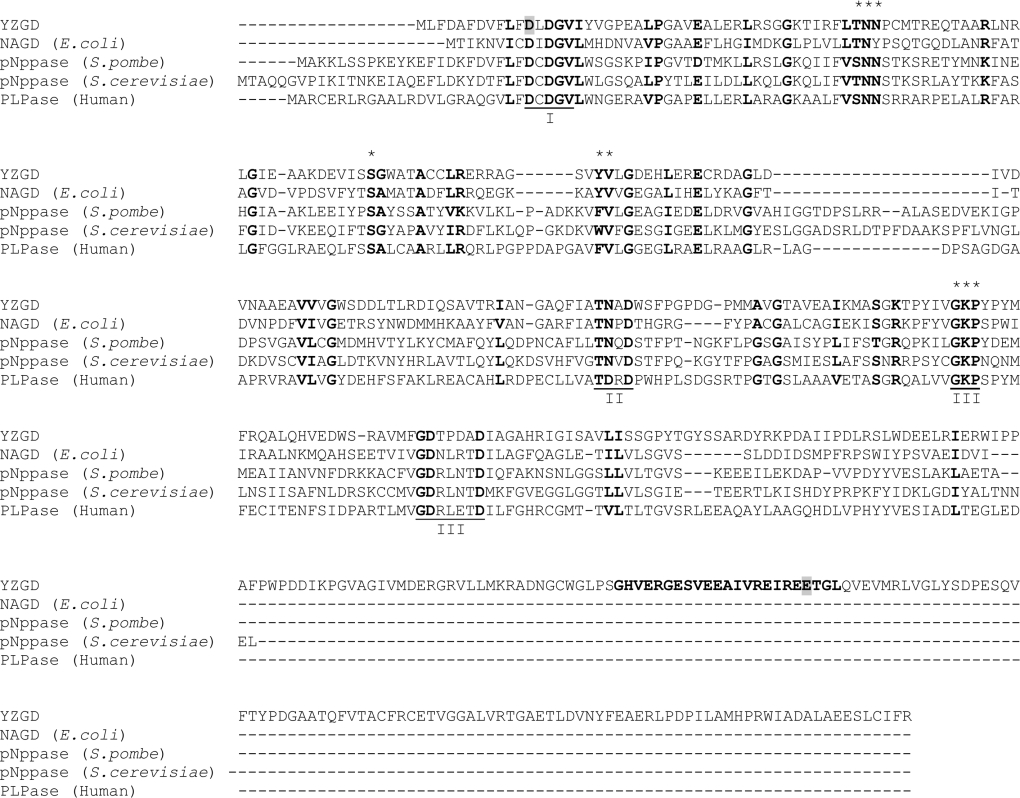
Figure 6. Alignment of YZGD with other members of the nitrophenyl phosphatase family.
The amino acid sequences of YZGD, NAGD from E. coli, the p-nitrophenyl phosphatases (pNppase) from S. pombe and S. cerevisiae and the human PLPase were aligned using CLUSTAL X [26]. Similar and identical amino acids are in boldface. Gaps in the amino acid sequences inserted to optimize alignment are indicated by a ‘-’. Motifs I, II and III of the HAD superfamily are underlined and labelled. The 23 amino acid Nudix signature sequence in YZGD is in boldface. The amino acids Asp12 (within the HAD domain of YZGD) and Glu324 (within the Nudix signature sequence of YZGD), which were modified by site-directed mutagenesis, are highlighted in grey. The [S/T]NN and [F/W/Y][I/L/V] motifs, which appear to be predictive markers for the nitrophenyl phosphatase family are denoted by a ‘*’, as are the conserved serine between motifs I and II and the conserved GKP sequence of motif III.