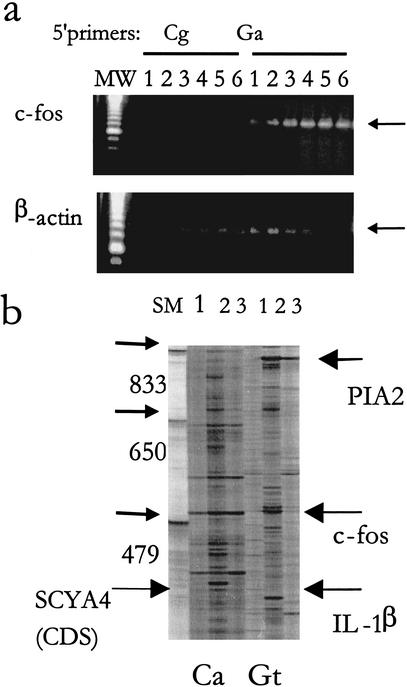
Figure 4.
Specificity of subset-specific 5′ primers. (a) Forty ng cDNA was used for the ARE-cDNA PCR with different initial annealing temperatures in the first four cycles (1, 32.5°C; 2, 35°C; 3, 37.5°C; 4, 40°C; 5, 42.5′°C; and 6, 45°C) followed by a consistent cycle of high annealing temperature (60°C). Two different 5' primers were used: Ga 5′ primer (which includes 10-mer sequence that targets a subset containing c-fos mRNA) and Cg 5′ primer as a control. Aliquots of the amplified ARE products were subjected to PCR specific to c-fos and β-actin. (b) Long Range display using two different primers showing two different patterns of display using 5′ primer Ca and Gt. cDNA samples (40 ng) from THP-1 cells (lane 1), from THP-1 cells that were treated with LPS+CHX (lane 2), or THP-1 cells that were treated with PMA (lane 3), were subjected to ARE-cDNA PCR at 42.5°C initial annealing temperature as described above. Samples were loaded on 4.5% urea-denaturing polyacrylamide gel and electrophoresed in the Genomyx LR sequencing apparatus for 16 hr. Arrows indicate differentially expressed bands that were upregulated by treatment of the LPS and CHX and with known identity. The full-length coding region of SCYA4 (CDS) is shown with large arrow. MW: molecular weight markers using amplified products of known sizes.