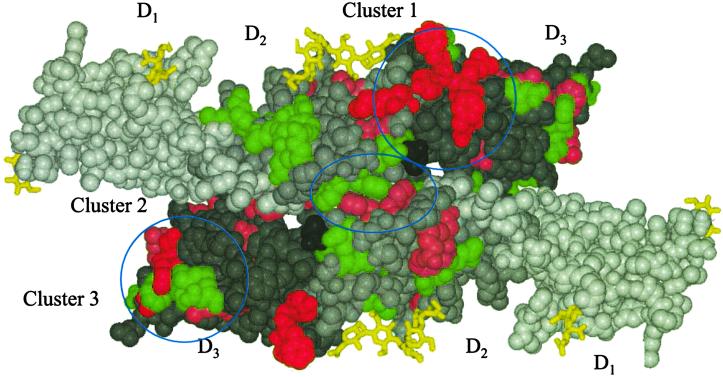
Fig 5.
A molscript (56) CPK representation of a crystallographic dimer of IL-6R, viewed toward the membrane anchors, showing the three mutational clusters found on the IL-6R structure. The crystallographic twofold axis lies at the center of the figure and perpendicular to the paper; the dimer interface lies east to west through the twofold axis. Residues in the upper molecule in the figure are colored to represent mutations that affect IL-6 binding to IL-6R (red, pink, and green spheres representing the atoms of residues that have <25%, 25–75%, and >75% of the binding activity of the wild type, respectively). Residues in the lower molecule are colored to represent mutations that affect gp130 signaling (red, pink, and green spheres representing the atoms of residues that have <25%, 25–75%, and >75% of the signaling of wild type, respectively). Residues in domains D1, D2, and D3 that have no mutational data or alter the structure are colored white, light gray, and gray, respectively. The cysteinyl-cysteine C192 is colored dark gray, and carbohydrate is represented by yellow bonds.