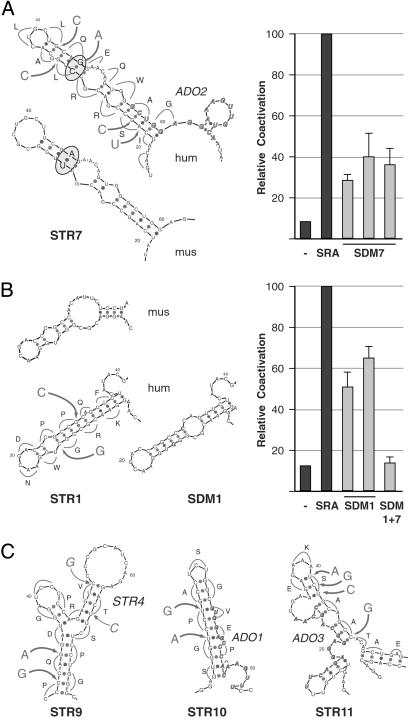
Fig 3.
Discrete RNA structure modeling and silent, site-directed mutagenesis. Representation of comparative low-resolution RNA modeling (STR) and silent mutagenesis strategy of discrete portions of SRA sequences from human (hum) and mouse (mus) as predicted by the mfold program (Left), and relative coactivation of corresponding human site-directed SRA mutants (SDMs) tested in tissue culture experiments (Right). The single-letter code of the presumptive amino acid sequence given by the ORF in SRA, nucleotide changes (large letters), and sites of mutations (arrows) are shown alongside the plotted representation of the inferred wild-type structure of the human SRA sequence. Bold, italic sequences in STR7, STR10, and STR11 annotate the complement sequences of ADOs (see Discussion). Relative transactivation of wild-type SRA (SRA) and site-directed mutants (SDM) were obtained by transient cotransfection of recombinant cDNAs (1–2 μg) or empty vector (−) along with PR expression vector (20 ng) and MMTV-Luciferase reporter (2.5 μg) into HeLa cells. Luciferase activities are shown as the mean (±SE) values of at least four separate experiments. (A) Site-directed mutations in predicted substructure 7. Covariation is indicated by the U-A pairing in mouse and C-G pairing in the human ortholog (circled). Three independent cDNA constructs of mutated STR7 (SDM7) were assayed in transient transfection experiments as described above. (B) Inferred STR1 located at the 5′ end of the SRA core sequence (mus, hum), targeting strategy (hum), and predicted structure after site-directed mutagenesis (SDM1). Two independent silent mutants were generated and tested. SDM1 + 7 is a SRA variant with silent mutations in both structure 1 and structure 7. (C) Predicted substructures 9–11, silent mutagenesis and target sequences for ADOs as indicated. Transactivation data are shown in Fig. 4.