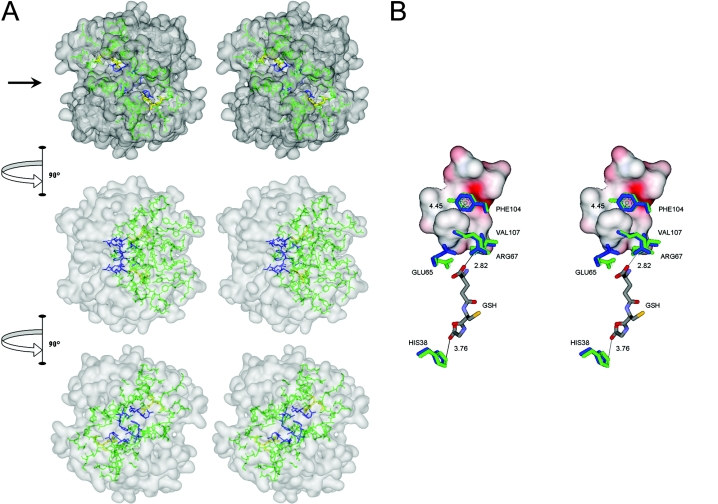
Figure 7. Interactions of amino acids in the lock-and-key ‘Clasp’ motif of adGSTD4-4.
(A) Stereo view of the lock-and-key ‘Clasp’ motif connecting two active site pockets of adGSTD4-4. The surface of the dimeric enzyme is shown in grey, GSH in each active site in yellow, the active- site amino acids in green and the Clasp residues in blue. Five of the six Clasp amino acids are also active-site residues. The top panel is looking down on to the 2-fold axis at the two active sites, which are diagonal to each other. The arrow in the top panel points to the interface groove that runs across the protein. The middle panel shows the GST rotated 90° in the direction indicated and the bottom panel shows the GST rotated a further 90° in the same direction. (B) Stereo view showing communication of GSH binding from one active site to the other through residues involved in the Clasp motif. Crystal structure of adGSTD4-4 (PDB id: 1JLW) is superimposed on adGSTD3-3 (PDB id: 1JLV) by fitting 1616 backbone atoms, with an R.M.S.D. (root mean square deviation) of 0.82 Å (1 Å=0.1 nm). Distances between centroids of aromatic ‘key’ residues are monitored from adGSTD4-4, whereas distances of GSH-interacting residues are monitored from the adGSTD3-3 enzyme complex. Amino acid labelling is for adGSTD4-4. Amino acid residues of adGSTD4-4 and adGSTD3-3 are coloured in blue and green respectively. The surface representation is of ‘lock’ residues from the subunit partner.