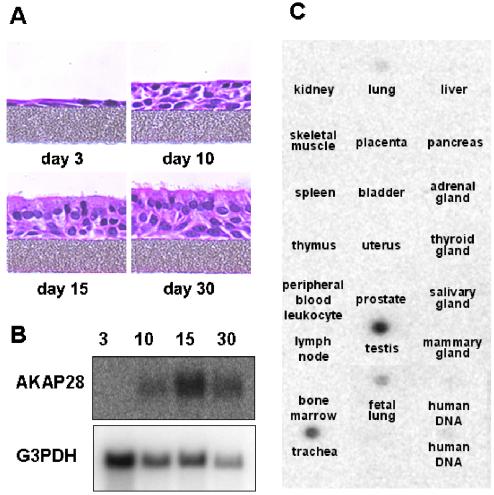
Figure 5.
Northern blot analysis of AKAP28. (A) Code-matched HBE cultures were differentiated on semipermeable supports. Cultures were fixed postplating at day 3, 10, 15, and 30. Cells were sectioned and stained with H&E. (B) Total RNA isolated from differentiating HBE cells at 3, 10, 15, and 30 d postplating (20 μg/lane for samples 3, 10, and 15; 6 μg/lane for 30) was analyzed by Northern blot with 32P-random primed cDNA corresponding to nucleotides 98–400 of AKAP28 cDNA. The same blot was probed with G3PDH as a loading control. Similar results were obtained on two separate blots. (C) A multiple tissue expression array was probed with the same AKAP28 probe used in B. A selected panel is shown; all tissues not shown were negative for the signal.