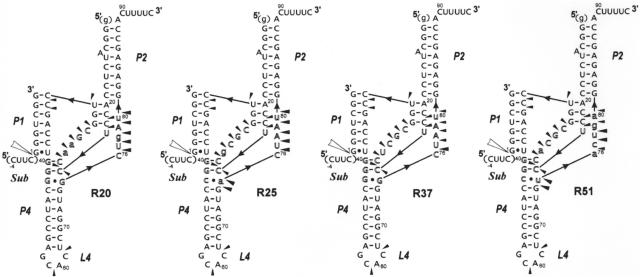
Figure 4.
Secondary structure models of trans-acting ribozymes: R20, R25, R37 and R51 and the results of their structural probing by Pb2+-induced cleavage method. The models derive from the consensus delta ribozyme pseudoknot structure. Nucleotides different than those found at corresponding positions of the wild-type sequence are marked by lower case letters. Outlined letters denote nucleotides present in the regions, which were randomized in the initial combinatorial library that was used in the selection of the ribozymes shown in the figure. Cleavages induced by Pb2+ ions are denoted by black triangles and the data derived from the autoradiograms which are shown in Supplementary Figure 1. The ribozyme components that were used for structural probing were deprived of the 5′-terminal guanosine residue in order to increase the homogeneity of RNA transcripts at their 5′ ends.