Abstract
Chlorophyll precursors, photosynthetic electron transport, and sugars have all been shown to be involved in signaling from the chloroplast to the nucleus, suggesting the presence of multiple signaling pathways of coordination between these two cellular compartments.
The endosymbiotic theory of chloroplast evolution proposes that a photosynthetic prokaryote was engulfed by a eukaryotic host to produce the eukaryotic plant cell. The ensuing endosymbiosis resulted in gene transfer from the chloroplast genome to the nuclear genome, and thus the chloroplast contains both nucleus-encoded and chloroplast-encoded components. The development of a fully functional chloroplast therefore depends on the coordinate expression of nuclear and chloroplast genes in response to both developmental and environmental signals. Chloroplast function requires the import of both nucleus-encoded photosynthetic proteins and cytoplasmic factors that regulate the expression of chloroplast genes. The plastid also plays a role in nuclear gene expression, with signals that originate in the chloroplast acting to regulate transcription of nucleus-encoded photosynthetic genes. In the last ten years, many studies have revealed the nature of nucleus-derived molecules that affect chloroplast gene expression at all levels (reviewed in [1,2,3]). Although it has been known for many years that the expression of a subset of nuclear genes, whose products are involved in photosynthesis, depends on the presence in the cell of functional plastids [4], little progress has been made in elucidating the signaling molecules or mechanisms involved in this retrograde signaling. Several recent discoveries have made inroads into this complex mechanism [5,6,7,8] and have begun to shed light on the black box of signaling from the chloroplast to the nucleus.
Chlorophyll precursors signal from the chloroplast
It has long been suspected that chlorophyll precursors play a role in signaling to the nucleus. Indirect evidence came from reports of the repression of nuclear photosynthetic genes in Chlamydomonas reinhardtii by the addition of a specific inhibitor of chlorophyll biosynthesis that results in chlorophyll-precursor accumulation [9,10]. The same chlorophyll precursors have also been reported to act as inducers of nucleus-encoded cytosolic and chloroplast-localized heat-shock proteins [11,12].
Two recent papers, by Mochizuki et al. [5] and Møller et al. [6], have identified proteins involved in chlorophyll biosynthesis as regulators of light-stimulated expression of nucleus-encoded photosynthetic genes such as those encoding the light-harvesting chlorophyll a/b binding protein (Lhcb), Rubisco small subunit (RbcS), chalcone synthase (Chs) and ferredoxin-NADP reductase (Fnr).
Mochizuki and co-workers examined Arabidopsis genomes-uncoupled (gun) mutants [13], in which Lhcb and RbcS transcription is derepressed in the absence of functional chloroplasts. Identification and analysis of several such gun mutants suggested the existence of multiple signaling pathways. Cloning of the GUN5 locus identified its product as the ChlH subunit of Mg-chelatase, a protein involved in chlorophyll synthesis, again suggesting that chlorophyll precursors act as a signal in one of these pathways. As shown in Figure 1, Mg-chelatase is required for the production of Mg-protoporphyrin IX monomethyl ester (MgProtoME). Inhibition of Lhcb gene expression in response to MgProtoME accumulation [9], and a requirement for cytoplasmic MgProtoME accumulation for the expression of Hsp70a and Hsp70b [12], suggests that this precursor may act as a signaling molecule between the chloroplast and the nucleus. But Mochizuki et al. show that two other mutants that disrupt the ChlI subunit (and thus the activity of Mg-chelatase) and have similarly pale chlorophyll phenotypes to gun5, do not show derepression of Lhcb and RbcS transcription. These data indicate that the ChlH subunit itself is instrumental in providing the signal from the chloroplast that regulates the expression of nuclear photosynthetic genes, perhaps in response to changes in the flux of chlorophyll biosynthetic precursors such as MgProtoME.
Figure 1.
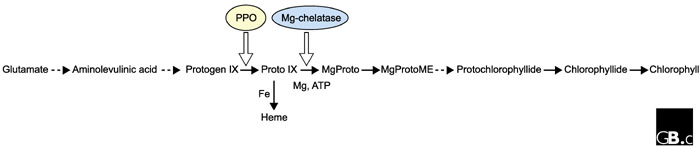
The pathway of chlorophyll biosynthesis. Solid arrows represent single steps; dashed arrows represent multiple steps; open arrows represent catalysis by an enzyme. Abbreviations: protogen IX, protoporphyrinogen IX; proto IX, protoporphyrin IX; MgProto, Mg-protoporphyrin IX; MgProtoME, Mg-protoporphyrin monomethyl ester; PPO, protoporphyrinogen IX oxidase.
Møller et al. isolated Arabidopsis long after far red (laf) mutants displaying reduced hypocotyl-growth inhibition in response to far-red light. These mutants also have an inability to become green following treatment with far-red light, and they have reduced expression of the light-regulated genes Lhcb, Chs, and Fnr in far-red light. LAF6 was cloned and shown to encode a novel plant member of the small soluble ATP-binding-cassette transporter family (atABC1); members of this family are usually involved in the import of catabolites across membranes. AtABC1 has a functional amino-terminal chloroplast-transit peptide and localizes to the periphery of chloroplasts, consistent with having a position at the inner envelope. The pale-green phenotype of laf6 seedlings suggested a deficiency in chlorophyll biosynthesis. Subsequent analysis confirmed that laf6 mutants accumulate twice as much protoporphyrin IX (proto IX; see Figure 1) and 40% less chlorophyll than wild-type seedlings.
If the accumulation of proto IX was the cause of the laf phenotype, treatment of seedlings with a herbicide that gives rise to artificially high cytoplasmic proto IX levels should result in the laf phenotype. This indeed proved to be the case, as treatment of wild-type seedlings with flumioxazin, an inhibitor of protoporphyrinogen IX oxidase (PPO), resulted in the same phenotype as laf6. Overexpression of atABC1 reduced the sensitivity of the seedlings to flumioxazin, as predicted. Given that PPO oxidizes protoporphyrinogen IX to proto IX in the chloroplast envelope (see Figure 2), these results suggest that atABC1 is responsible for the import of proto IX into the stroma. In the absence of atABC1 in the laf6 mutant, proto IX is not imported and it therefore accumulates in the cytoplasm, resulting in repression of the expression of Lhcb, Chs and Fnr. Both of these recent studies [5,6] show the importance of the chlorophyll biosynthetic pathway in chloroplast development and in signaling between the chloroplast and nucleus.
Figure 2.
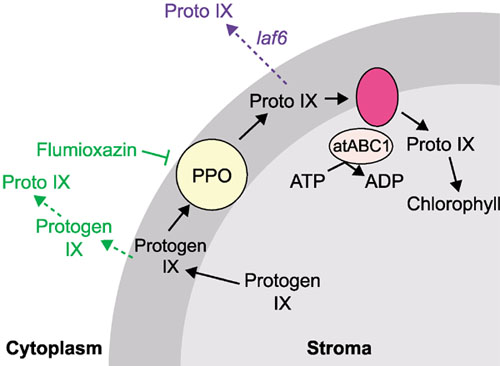
A model of the role of atABC1 and PPO in chlorophyll biosynthesis. PPO oxidizes protogen IX to proto IX in the chloroplast envelope (dark gray), whereupon it is imported back into the stroma by atABC1 and an integral membrane protein. In the laf6 mutant (shown in blue), proto IX import is inhibited, allowing its accumulation in the cytoplasm. In the presence of flumioxazin (shown in green), PPO is inhibited, allowing protogen IX into the cytoplasm where it is oxidized non-specifically to proto IX, thus mimicking the laf6 phenotype. Adapted from [6]; for abbreviations, see Figure 1 legend.
Plastid redox state and sugars are involved in signaling from the chloroplast
The expression of nucleus-encoded photosynthetic genes, such as Lhcb, has been shown to be repressed in the presence of high levels of sugar (reviewed in [14]). It has also been proposed that photosynthetic electron transport is required to activate Lhcb transcription [15]. Oswald et al. [7] investigated whether these two effects operate through a common signaling pathway. They showed that photosynthetic electron transport is essential for the generation of a chloroplast signal that activates transcription of Lhcb and RbcS. This transcriptional activation did not depend on sugar status, implying that it is an independent event. Sucrose feeding repressed the transcription of Lhcb and RbcS, however, suggesting that the two pathways may interact in controlling the expression of these nuclear genes.
In studies of a mutant of the maize Sucrose export defective (Sxd1) gene, Provencher et al. [8] reported accumulation of sugars in photosynthetic bundle sheath cells as a result of a defect in sucrose export to the vascular parenchyma cells. The sxd1 mutant did not show repression of the nucleus-encoded photosynthetic genes Lhcb and RbcS, despite accumulation of high sugar levels. This result implies that SXD1 has a role in repression of Lhcb and RbcS in response to high sugar levels. On the basis of this observation and the localization of Sxd1 to the chloroplast, the authors proposed that SXD1 might play a role in the sugar-sensing mechanism involved in chloroplast-to-nucleus signaling.
A model of chloroplast-to-nucleus signaling
The four papers described above [5,6,7,8] have provided evidence for the involvement in signaling between the chloroplast and nucleus of components of the chlorophyll biosynthetic pathway, the redox state of the chloroplast, and sugars. It is clear that there are multiple signaling pathways that result in repression or stimulation of the same set of nuclear genes in response to different cellular or environmental cues. These recent discoveries call for a revision of the model proposed to describe signaling from the chloroplast to the nucleus (Figure 3). Chlorophyll precursors, such as MgProtoME and proto IX, accumulate in response to reduced rates of chlorophyll synthesis. These precursors may enter the cytoplasm and bind to factors that signal the repression of expression of nucleus-encoded photosynthetic genes. Alternatively, they may bind to Mg-chelatase at the chloroplast inner envelope, upon which the Mg-chelatase would send a signal that resulted in the repression of transcription. It has been shown that chloroplast electron transport is required for the activation of expression of nucleus-encoded photosynthetic genes, and the redox-sensitive molecules thioredoxin or glutathione may be involved in this signal transduction, possibly through an as-yet unidentified inner-envelope component. Sugars also affect expression of nucleus-encoded photosynthetic genes and may act via Sxd1 in the chloroplast or hexokinase in the cytoplasm [14], sending a signal to repress the expression of photosynthetic genes when a source of carbon is abundant. It has also been proposed that cytoplasmic kinases and phosphatases may play a role in signaling from the chloroplast to the nucleus [15,16,17]. Several of the signaling pathways described above may use common kinases and phosphatases as transducers of the signal, a frequent occurrence in eukaryotic cells.
Figure 3.
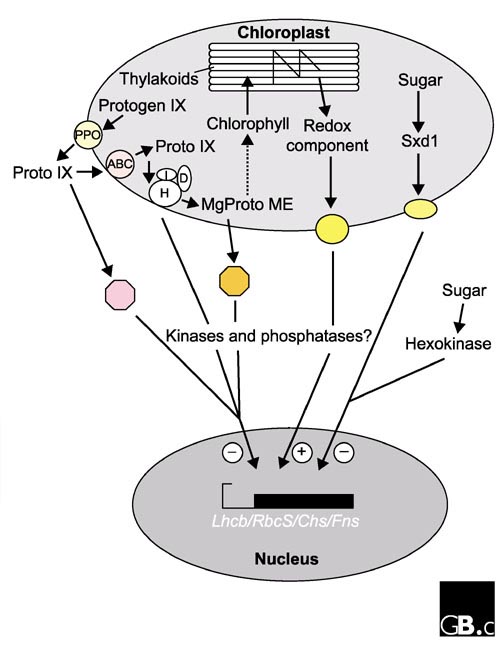
A summary of the pathways of chloroplast-nucleus signaling proposed in this article. H, I and D are the ChlH, ChlI and ChlD subunits of Mg-chelatase, respectively; the octagons represent possible receptors for proto IX (pink) and MgProtoME (orange), respectively; the yellow ovals represent unidentified chloroplast envelope proteins involved in signal transduction. Plus and minus signs denote a positive or negative effect on transcription in the nucleus, respectively.
Undoubtedly, there is still much to be discovered in this complex field. It will become increasingly difficult to study individual pathways in isolation, as one can envisage an elaborate network of crosstalk between the pathways. As these recent papers have shown, however, the search for retrograde signaling components that pass from the plastid back to the nucleus is gathering momentum.
Acknowledgments
Acknowledgements
Work in our laboratory is supported by funds from the US Department of Energy (93ER70116) and the National Institutes of Health (GM54659) to S.P.M.; E.C.B. and A.S. are supported by Skaggs post-doctoral fellowships.
References
- Somanchi A, Mayfield SP. Nuclear-chloroplast signalling. Curr Opin Plant Biol. 1999;2:404–409. doi: 10.1016/s1369-5266(99)00013-8. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Coordination of nuclear and chloroplast gene expression in plant cells. Int Rev Cytol. 1998;177:115–180. doi: 10.1016/s0074-7696(08)62232-9. [DOI] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. doi: 10.1016/S0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- Mayfield SP, Taylor WC. Carotenoid-deficient maize seedlings fail to accumulate light-harvesting chlorophyll a/b binding protein (LHCP) mRNA. Eur J Biochem. 1984;144:79–84. doi: 10.1111/j.1432-1033.1984.tb08433.x. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller SG, Kunkel T, Chua N-H. A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev. 2001;15:90–103. doi: 10.1101/gad.850101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald O, Martin T, Dominy PJ, Graham IA. Plastid redox state and sugars: Interactive regulators of nuclear-encoded photosynthetic gene expression. Proc Natl Acad Sci USA. 2001;98:2047–2052. doi: 10.1073/pnas.021449998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencher LM, Miao L, Sinha N, Lucas WJ. Sucrose export defective 1 encodes a novel protein implicated in chloroplast-to-nucleus signaling. Plant Cell. 2001;13:1127–1141. doi: 10.1105/tpc.13.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanningmeier U, Howell SH. Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardtii. J Biol Chem. 1984;259:13541–13549. [PubMed] [Google Scholar]
- Johanningmeier U. Possible control of transcript levels by chlorophyll precursors in Chlamydomonas. Eur J Biochem. 1988;177:417–424. doi: 10.1111/j.1432-1033.1988.tb14391.x. [DOI] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rudiger W, Beck CF. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc Natl Acad Sci USA. 1997;94:14168–14172. doi: 10.1073/pnas.94.25.14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J, Oster U, Rudiger W, Beck CF. Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 2000;24:523–531. doi: 10.1046/j.1365-313X.2000.00898.x. [DOI] [PubMed] [Google Scholar]
- Susek R, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Jang JC, Sheen J. Sugar sensing in higher plants. Plant Cell. 1994;6:1665–1679. doi: 10.1105/tpc.6.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas J-M, Lomas M, LaRoche J, Falkowski PG. Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Protein phosphatase activity is required for light-inducible gene expression in maize. EMBO J. 1993;12:3497–3505. doi: 10.1002/j.1460-2075.1993.tb06024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandok MR, Sopory SK, Oelmuller R. Cytoplasmic kinase and phosphatase activities can induce PsaF gene expression in the absence of functional plastids: evidence that phosphorylation/dephosphorylation events are involved in interorganellar crosstalk. Mol Gen Genet. 2001;264:819–826. doi: 10.1007/s004380000371. [DOI] [PubMed] [Google Scholar]


