Figure 2.
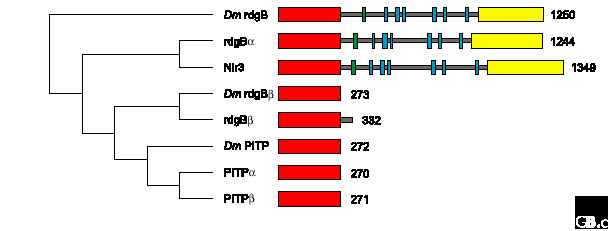
Structural relationships within the PITP family. A ClustalW alignment of human and Drosophila (Dm) PITP sequences was made using MacVector (version 7.0, Oxford Molecular). A bootstrapped dendrogram (1,000 replications) was constructed by rooting with the Drosophila rdgB sequence, using uncorrected, neighbor-joining parameters and ignoring gap sites. The corresponding domain topologies on the right illustrate the number of amino-acid residues in each protein and the position of PITP domains (red), Ca2+-binding domains (green), short hydrophobic regions (blue) and Pyk2-binding domains (yellow).
