Abstract
We identified a novel gene ST13 from a subtractive cDNA library of normal intestinal mucosa in 1993, more studies showed that ST13 was a co-chaperone of Hsp70s. Recently we detected the ST13 gene expression in tumor tissue and adjacent normal tissue of the same colorectal cancer patient and investigated if the ST13 gene expression might have any prognostic value. Analysis was performed at molecular level by reverse transcription-PCR using real-time detection method. We measured two genes simultaneously, ST13 as the target gene and glyceraldehydes-3-phosphate dehydrogenase as a reference gene, in primary colorectal tumor specimens and tumor-adjacent normal mucosa specimens from 50 colorectal cancer patients. The expression levels of the ST13 gene were significantly decreased in primary tumors compared with adjacent mucosa (P<0.05). But there were no significant differences in the expression of ST13 as compared with different Dukes’ stage, tumor differentiation grade, invasion depth, lymph node metastasis and disease-specific survival.
Keywords: ST13, Colorectal cancer, Real-time PCR
INTRODUCTION
Today, colorectal cancer (CRC) is the third leading cause of death due to cancer and the third most common cancer expected to occur in men and women (Jemal et al., 2004). Colorectal cancer development and metastasis are associated with altered gene expression profiles (Vogelstein and Fearon, 1988). We reported in 1993 the identification of a novel gene ST13 (HSU17714) from a subtractive cDNA library of normal intestinal mucosa, whose expression was down-regulated in colorectal cancer. Further studies showed that ST13 genomic DNA had 32017 base pairs, comprised of 12 exons. Correspondingly its cDNA had 3127 base pairs, the open reading frame (ORF) was 1170 base pairs and codes for a 369-amino acid protein. The gene was mapped to chromosome 22q13 (Zheng, 1993; 1997; Zheng et al., 1997).
Hohfeld et al.(1995) reported an Hsc70-inter-acting protein (Hip) which bound the ATPase domain of Hsc70 in an ADP-dependent manner. P48 protein was identified as a transient component during the cell-free assembly of progesterone receptor complex in 1996 (Prapapanich et al., 1996a; 1996b). ST13 showed >90% identity at the amino acid level with the rat Hip and P48. More studies showed that Hip/P48/ST13 was associated with Hsp70 and Hsc70, affected the Hsc70/Hsp70 chaperone activities in vivo and in vitro (Nollen et al., 2001; Fan et al., 2002; Irmen and Hohfeld, 1997). Molecular chaperones of the Hsp70 family play a key role in the control of protein folding during protein biogenesis, protein transport through membranes, and when cells are exposed to proteotoxic stress (Hartl, 1996; Bukau and Horwich, 1998). Our studies showed that ST13 might play a role in carcinogenesis and participate in regulating apoptosis (data not published).
Northern blot and RNA dot blot analyses showed that the expression level of ST13 gene was significantly lower in 15 cases of colorectal cancer than that in adjacent colonic mucosa. RNA dot blot analyses also showed that the ratio of low ST13 expression in colorectal carcinoma was significantly higher in lymph node metastasis patients compared with that in mucosa (Mo et al., 1996). In situ hybridization analyses showed that the ratio of low ST13 expression in colorectal carcinoma seemed higher in Duke’s C or lymph node metastasis patients compared with that in mucosa, but there was no significant difference (Ye et al., 2001). To our knowledge, except for the Duke’s stage and lymph node metastasis, the relationship between ST13 expression and other clinicopathological factors had not been studied.
Recent reports on real-time RT-PCR showed the advantages of this method compared with conventional methods (Gelmini et al., 1997; Swan et al., 1997). This method offers the possibility to perform multiplex PCR, simultaneous detection of the expression of more than one gene in the same tube, using specific sets of primers and specific probes with different fluorogenic labels for each gene to be investigated. We use this multiplex PCR for detection of expression of our target gene ST13 and the reference gene GAPDH. Measured the expression of the reference gene provides information on the integrity and amount of mRNA present in the tested samples. To clarify these disparate findings, we investigated if the expression of ST13 might have any clinic significance or prognostic value in this study.
MATERIALS AND METHODS
Samples
Tumor and paired mucosa samples were obtained from 50 colorectal carcinoma patients undergoing primary tumor resection at the Second Affiliated Hospital of Zhejiang University during the period between Sept. 2000 to May 2001. The ethics committee of Zhejiang University approved the study. Adjacent, normal-appearing mucosa were taken at a distance of ~10 cm from the tumors. The biopsies were snap-frozen in liquid nitrogen and stored at −80 °C until used. Surgical and pathological records were reviewed for Dukes’ stage, tumor differentiation grade, age, gender, and tumor localization. The growth pattern and the grade of differentiation were classified by pathologists as recommended by the WHO. The malignant tumors were classified according to Dukes’ stage, as modified by others, into the pathological stages Dukes’ A (TNM stage I), Dukes’ B (TNM stage II), Dukes’ C (TNM stage III), and Dukes’ D (TNM stage IV). Clinical characteristics of the patients are presented in Table 1.
Table 1.
Clinical characteristics of the patients
| Variable | Number of patients (%) |
| All patients | 50 |
| Gender | |
| Male | 26 (52) |
| Female | 24 (48) |
| Tumor site | |
| Colon | 26 (52) |
| Rectum | 24 (48) |
| Dukes’ stage | |
| A | 8 (16) |
| B | 19 (38) |
| C | 21 (42) |
| D | 2 (4) |
| Histology | |
| Well differentiated | 14 (28) |
| Moderately differentiated | 20 (40) |
| Poorly differentiated | 3 (6) |
| Mucinous adenocarcinoma | 13 (26) |
Total RNA extraction and reverse transcription of mRNA
Isolate total RNA from TriZol (Invitrogen). Adjust concentration of total RNA to 0.2 μg/μl after RNA isolation. Take 10 μl total RNA, add 1 μl oligo (dT) (Promega), 70 °C for 10 min, put on ice for 2 min, then add 4 μl 5×1st strand buffer, 2 μl 0.1 mol/L DTT, 1 μl 10 mmol/L each dNTP (Gibcol), and 1 μl RNase inhibitor, mix well, 42 °C for 2 min. Add 1 μl SSRTII (Gibcol) to each tube, 42 °C for 50 min, then 70 °C for 15 min. Spin reagent down, add 80 μl pure water, mix well, spin down, store at −20 °C for real time PCR.
Real-time quantitative PCR
Quantitative PCR was performed using the ABI PRISM 7700 Sequence Detector (Applied Biosystems). The house keeping gene GAPDH was used as endogenous control to compensate for the variation in RNA amount and to check the efficiency of the reverse transcription reaction. PCR samples were prepared as follows: 2 μl of cDNA were transferred into MicroAmp Reaction Tubes already containing 48 μl of reaction mixture. The components of the reaction mixture contained 25 μl 2×universal buffer (Roche), 6 μl target ST13 gene primer-probe mix, 2.5 μl human GAPDH primer-probe mix, 14.5 μl H2O. The condition for the PCR were 2 min at 50 °C and 10 min at 95 °C, cycling parameters were 15 s at 95 °C and 1 min at 60 °C (40 cycle), hold at 15 °C. Sequences of the used primers and probes are listed in Table 2. All of the samples were amplified simultaneously in triplicate in a one assay-run. The quantitative data were calculated according to the instructions given by Applied Biosystems.
Table 2.
DNA sequences of the primers and probes used in the PCR
| Gene | Primer/probe |
| GAPDH | Forward primer: 5′-CTTAGCACCCCTCCCCAAG-3′ |
| Reverse primer: 5′-GATGTTCTGGAGAGCCCCG-3′ | |
| Probe: 5′-(VIC)CATGCCATCACTGCCACCCAGAAGA(TAMRA)-3′ | |
| ST13 | Forward primer: 5′-CCTCGCTTGGCCATTTTGT-3′ |
| Reverse primer: 5′-TGGCAGCATTTGGCTTCTG-3′ | |
| Probe: 5′-(FAM)CCAAGAGGGCCAGTGTCTTCGTCAAA(TAMRA)-3′ |
Statistical analysis
The clinicopathological variables used in this study were as follows: Dukes’ stage, differentiation grade, lymph node metastasis, and invasion depth. The obtained data were analyzed by statistical modelling using commercial software SPSS. Unless otherwise stated, the data were presented as means and SDs. To compare sets of continuous parameters measured in the same tumor tissues, Spearman’s correlation coefficient® and the Wilcoxon signed-rank test were used. The statistical significance of the difference in survival of the groups was calculated using log-rank test. Relative risk was assessed by univariate and multivariate Cox proportional hazard model. Statistical values of P≤0.05 were considered to be significant.
RESULTS
Clinical characteristics of the patients
As shown in Table 1, the median age of the patients was 60 (range, 25~91) years. Twenty-six patients were male, and 24 were female. Among 50 patients, 26 patients had colon cancer, and 24 had rectal carcinoma. Of the primary carcinomas, 14 were highly, 20 were moderately, and 3 were poorly differentiated, 13 were mucinous adenocarcinoma. Primary tumor stage was Dukes’ A in 8 patients, B in 19 patients, C in 21 patients, and D in 2 patients. At the end of the study, we could follow up 22 patients. The median following-up time was 50 months.
Gene expression levels in colorectal mucosa and carcinomas
The relative gene expression levels of ST13 in the mucosa and in carcinomas are presented in Table 3. As shown, significantly lower expression levels were found in tumors as compared with mucosa (t=2.547, P=0.014). The ratio of low expression (ST13 expression in colorectal carcinoma/colorectal mucosa≤1) samples was 62% (31/50).
Table 3.
ST13 expression in matched samples of normal colorectal mucosa (N) and primary colorectal cancer (T)
| Cases | Min | Max | x¯±s | |
| T | 50 | −3.56 | 9.5 | 5.1470±2.1280 |
| N | 50 | −3.20 | 7.4 | 4.6653±1.7147 |
Gene expression levels in different Dukes’ stage CRC patients
Analyses showed that there was no significant difference in different Dukes’ stage group (F=1.329, P=0.227) in Table 4.
Table 4.
ST13 expression in different Dukes’ stage CRC patients
| Dukes’ stage | Cases | x¯±s | F value | P value | Low expression samples (%) |
| A | 8 | 0.9977±0.7604 | 1.329 | 0.227 | 5 (62.5) |
| B | 19 | 1.2949±1.1309 | 11 (57.9) | ||
| C | 21 | 0.7537±0.5307 | 14 (66.7) | ||
| D | 2 | 1.1230±0.2778 | 1 (50) |
Gene expression levels in different differentiation stage CRC patients
As shown in Table 5, there was no significant difference in different differentiation stage (F=1.525, P=0.221).
Table 5.
ST13 expression in different differentiation grade CRC patients
| Differentiation grade | Cases | x¯±s | F value | P value | Low expression samples (%) |
| Well differentiated | 14 | 0.7454±0.4618 | 1.525 | 0.221 | 10 (71.4) |
| Moderately differentiated | 20 | 1.3240±1.0983 | 10 (50) | ||
| Poorly differentiated | 3 | 0.8332±0.9181 | 2 (66.7) | ||
| Mucinous adenocarcinoma | 13 | 0.8740±0.6560 | 9 (69.2) |
Gene expression levels in different invasion depth CRC patients
As shown in Table 6, there was no significant difference in different invasion depth groups (F=0.248, P=0.782).
Table 6.
ST13 expression in different invasion depth CRC patients
| Invasion depth | Cases | x¯±s | F value | P value | Low expression samples (%) |
| T1 | 2 | 1.4062±0.6873 | 0.248 | 0.782 | 1 (50) |
| T2 | 13 | 0.9425±0.8365 | 9 (69.2) | ||
| T3 | 35 | 1.0247±0.8898 | 21 (60) |
Gene expression levels in different lymph node metastasis CRC patients
As shown in Table 7, there was no significant difference in positive and negative lymph node metastasis group (F=1.848, P=0.072).
Table 7.
ST13 expression in negative and positive lymph node metastasis CRC patients
| Lymph node metastasis | Samples | x¯±s | F value | P value | Low expression samples (%) |
| Negative | 27 | 1.2068±1.0296 | 1.848 | 0.072 | 16 (59.2) |
| Positive | 23 | 0.7873±0.5199 | 15 (65.3) |
Relationship between ST13 expression and survival
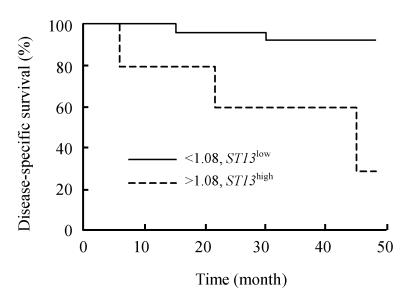
Among the 50 CRC patients, we could conduct 22 following-up investigations. Primary tumor stage was Dukes’ A in 4 patients, B in 7 patients, C in 9 patients, and D in 2 patients. The ratio of low expression cases was 68.2% (15/22). As expected, 4-year tumor-specific survival of the patients gradually decreased from those classified as Duke’s A (100%), Duke’s BC (75%), Duke’s D (0%). As shown in Fig.1, the 4-year survival rate was 80% (12/15) for patients of group ST13 low and 57.2% (4/7) for patients of group ST13 high. But there was no significant difference between them (P=0.209).
Fig. 1.
Kaplan-Meier curves for disease-specific survival in 22 CRC patients
ST13 expression in colorectal carcinoma/colorectal mucosa≤1.08: —ST13 low; ST13 expression in colorectal carcinoma/colorectal mucosa>1.08: ---- ST13 high
DISCUSSION
Hip/ST13 cooperates with Hsp70 in protein folding by stabilizing the ADP-bound state of Hsp70. Besides affecting the Hsp70 chaperone cycle, Hip/ST13 alone can also bind to unfolded proteins and prevent their aggregation, but refolding of protein to their active requires cooperation with other chaperones (Bruce and Churchich, 1997). Other classes of Hsp70 co-chaperones include BAG-1 (bcl-2 associated anti-death gene 1 protein) and CHIP (carboxyl terminus of Hsc70-interacting protein) cofactors (Hohfeld and Jentsch, 1997; Ballinger et al., 1999).
Hsp70 is molecular chaperone involved in many important biological processes. Abrogation of Hsp70 expression can induce apoptosis in tumor cells (Nylandsted et al., 2000; Ravagnan et al., 2001). Some studies showed that increased Hsp70-positive expression correlated significantly with low differentiation and was associated with worse overall survival in a series of 128 CRC patients (Lazaris et al., 1995), mitochondria Hsp70 (mortalin) over-expression correlated with poor survival in colorectal cancer patients (Dundas et al., 2005). BAG-1 is a recently identified Bcl-2 interacting anti-apoptotic protein. The percentage of CRC cases exhibiting nuclear BAG-1 positivity was significantly higher in distant metastasis-positive cases than in distant metastasis-negative cases, overall survival was significantly shorter for patients with tumors exhibiting BAG-1 positive nuclei than those without nuclear BAG-1-staining, and indicated that nuclear BAG-1 expression was the only independent prognostic variable for mortality (Kikuchi et al., 2002).
The real-time PCR result of the present study showed that the mean gene expression level of ST13 was significantly low in the tumor compared with the mucosa, and that the low expression ratio was 62% in 50 CRC patients. The result was in accordance with several studies using northern blot and RNA dot blot methods which showed the low expression ratio was 60% (Mo at al., 1996). Our study also showed that the expression level of ST13 gene in different clinicpathological factors group including Dukes’ stage, differentiation grade, lymph node metastasis, and invasion depth had no significant difference. The result supported the in situ histochemistry study (Ye et al., 2001). In our study, patients with high level of ST13 seemed to have poorer outcome (3 died in 7 cases) compared with patients having low level of ST13 (3 died in 15 cases). But there was no significant difference between them, because we could only follow-up 22 CRC patients. Maybe we can find the expression level of ST13 has some prognostic value if we get more patients involved in the study.
Footnotes
Project (Nos. 30400521 and 39770818) supported by the National Natural Science Foundation of China
References
- 1.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruce BD, Churchich J. Characterization of the molecular-chaperone function of the heat-shock-cognate-70-interacting protein. Eur J Biochem. 1997;245:738–744. doi: 10.1111/j.1432-1033.1997.00738.x. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/S0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 4.Dundas SR, Lawrie LC, Rooney PH, Murray JI. Mortalin is over-expressed by colorectal adenocarcinomas and correlates with poor survival. J Pathol. 2005;205(1):74–81. doi: 10.1002/path.1672. [DOI] [PubMed] [Google Scholar]
- 5.Fan JH, Yang W, Sai J, Richmond A. Hsc/Hsp70 interacting protein (Hip) associates with CXCR2 and regulates the receptor signaling and trafficking. J Bio Chem. 2002;277(8):6590–6597. doi: 10.1074/jbc.M110588200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelmini S, Orlando C, Sestini R, Vona J, Pinzani P, Giacca M, Pazzagali M. Quantitative polymerase chain reaction-based homogeneous assay with fluorogenic probe to measure c-erB-2 oncogene amplification. Clinical Chemistry. 1997;43(5):752–758. [PubMed] [Google Scholar]
- 7.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 8.Hohfeld J, Jentsch S. GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hohfeld J, Minami Y, Hartl FU. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell. 1995;83:589–598. doi: 10.1016/0092-8674(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 10.Irmen H, Hohfeld J. Characterization of functional domains of the eukaryotic co-chaperone Hip. J Bio Chem. 1997;272(4):2230–2235. doi: 10.1074/jbc.272.4.2230. [DOI] [PubMed] [Google Scholar]
- 11.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. American cancer society. Cancer statistics. CA Cancer J Clin. 2004;54(1):8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 12.Kikuchi R, Noguchi T, Takeno S, Funada Y, Moriyama H, Uchida Y. Nuclear BAG-1 expression reflects malignant potential in colorectal carcinomas. Br J Cancer. 2002;87(10):1136–1139. doi: 10.1038/sj.bjc.6600579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazaris AC, Theodoropoulos GE, Davaris PS, Panoussopoulos D, Nakopoulous Z, Ittas C, Golematis BC. Heat shock protein 70 and HLA-DR molecules tissue expression. Prognostic implications in colorectal cancer. Dis Colon Rectum. 1995;38(7):739–745. doi: 10.1007/BF02048033. [DOI] [PubMed] [Google Scholar]
- 14.Mo YQ, Zheng S, Shen DJ. Differential expression of HSU17714 gene in colorectal cancer and normal colonic mucosa. Chinese Journal of Oncology. 1996;18(4):241–243. (in Chinese) [PubMed] [Google Scholar]
- 15.Nollen EA, Kabakov AE, Brunsting JF, Kanon B, Hohfeld J, Kampinga HH. Modulation of in vivo Hsp70 chaperone activity by Hip and BAG-1. J Biol Chem. 2001;276:4677–4682. doi: 10.1074/jbc.M009745200. [DOI] [PubMed] [Google Scholar]
- 16.Nylandsted J, Rohde M, Brand K, Bastholm L, Elling F, Jaattela M. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspase and bypass of Bcl-2. Proc Natl Aca Sci USA. 2000;97:7871–7876. doi: 10.1073/pnas.97.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prapapanich V, Chen S, Nair SC, Rimerman RA, Smith DF. Molecular cloning of human P48, a transient component of progesterone receptor complexes and an Hsp70-binding protein. Molecular Endocrinology. 1996;10(4):420–431. doi: 10.1210/me.10.4.420. [DOI] [PubMed] [Google Scholar]
- 18.Prapapanich V, Chen S, Toran EG, Rimerman RA, Smith DF. Mutational analysis of the hsp70-interacting protein Hip. Molecular and Cellular Biology. 1996;16(1):6200–6207. doi: 10.1128/mcb.16.11.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravagnan L, Gurbuxani S, Susin SA, Maisse C, Daugas E, Zamzami N, Mak T, Jaattela M, Penninger JM, Garrido C, et al. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nature-Cell Biol. 2001;3:839–843. doi: 10.1038/ncb0901-839. [DOI] [PubMed] [Google Scholar]
- 20.Swan DC, Tucker RA, Holloway BP, Icenogle JP. A sensitive, type-specific, fluorogenic probe assay for detection of human papillomavirus DNA. J Clin Microbiol. 1997;35(4):886–891. doi: 10.1128/jcm.35.4.886-891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogelstein B, Fearon ER. Genetic alterations during colorectal tumor development. N Engl J Med. 1988;312(9):525. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 22.Ye F, Fang SC, Zheng S. In situ analysis the expression of new ST13 gene in colorectal cancer and its prognostic value. Journal of Practical Oncology. 2001;16(5):318–321. (in Chinese) [Google Scholar]
- 23.Zheng S. Application of subtractive hybridization in screening for colorectal cancer-related genes. Chin Med J. 1993;8:100. [PubMed] [Google Scholar]
- 24.Zheng S. Recent study on colorectal cancer in China: early detection and novel related gene. Chin Med J. 1997;110(4):309–310. [PubMed] [Google Scholar]
- 25.Zheng S, Cai XH, Cao J, Geng LY. Screening and identification of down-regulated genes in colorectal carcinoma by subtractive hybridization: a method to identify putative tumor suppressor genes. Chin Med J. 1997;110(7):543–549. [PubMed] [Google Scholar]