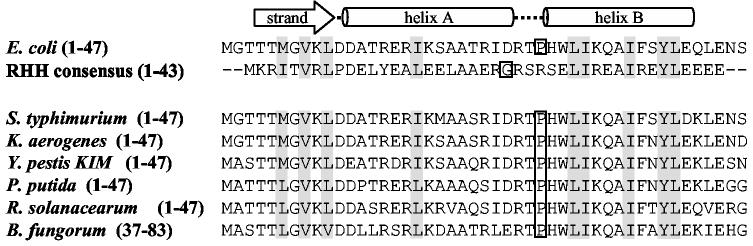
Fig. 3.
Predicted strand-helix-helix fold of PutA47 and sequence alignments. The secondary structure of PutA47 was predicted using the PSIPRED protein structure prediction server and is illustrated directly above the PutA47 amino acid sequence. The alignment of PutA47 with the RHH consensus sequence (RHH) of the CopG family is shown. A conserved domain search revealed the RHH fold at the N-terminal region of PutA from S. typhimurium (P10503), K. aerogenes (O52485), Y. pestis KIM (NP_669761), P. putida (AAF25000), R. solanacearum (NP_521420), and B. fungorum (ZP_00029196) as shown by the sequence alignments. Conserved hydrophobic residues are shaded in grey. The sequence alignments were obtained using the conserved domain architecture retrieval tool from NCBI.