Figure 3.
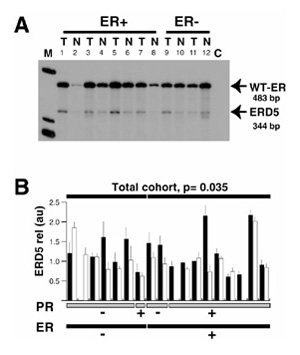
Comparison of the relative expression of exon 3 deleted estrogen receptor (ER) variant (ERD3) messenger RNA between breast tumor and adjacent matched normal breast samples. (A) Total RNA extracted from frozen tissue sections from tumor (T) and adjacent normal (N) breast tissue samples was reverse transcribed and polymerase chain reaction (PCR) amplified using D5U and D5L primers. Radioactive PCR products were separated on a 6% acrylamide gel and visualized by autoradiography. Bands that migrated at 483 and 344 base pairs were identified as corresponding to wild-type (WT)-ER and exon 5 deleted ER variant (ERD5) messenger RNA, respectively. C, negative control (no complementary DNA added during the PCR reaction). (B) For each case, signals corresponding to ERD5 variant messenger RNA were quantified and expressed in arbitrary units for tumor (black column) and normal (white column) components. For each sample, the mean and the standard deviation of at least three different PCR assays are indicated. Cases are sorted by ER status (black bottom lane) and progesterone receptor (PR) status (gray bottom lane). Samples that failed to have three measurable signals in the four experiments performed in both normal and neoplastic components were not included in the statistical analysis. The significance of the differences between tumor and normal matched components within each subgroup, as tested using the Wilcoxon matched-pair test, is indicated where P <0.05. m, molecular weight marker.
