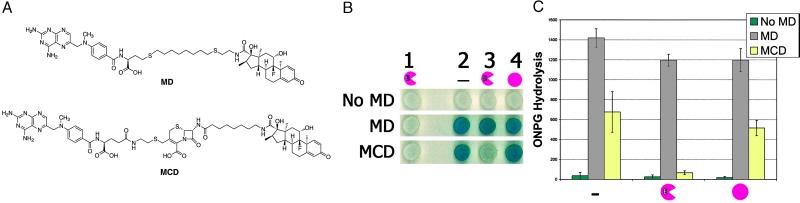
Fig 3.
Chemical complementation links enzyme catalysis to reporter gene transcription. (A) Structures of the Mtx-Dex (MD) and Mtx-Cephem-Dex (MCD) heterodimers. (B) X-Gal plate assays of cephalosporinase-dependent Mtx-Cephem-Dex-induced lacZ transcription. Yeast strains containing a lacZ reporter gene were grown on X-Gal indicator plates with or without Mtx-linker-Dex molecules as indicated. Columns 1–4 correspond to yeast strains containing a LexA DNA-binding domain fusion protein, a B42 activation domain fusion protein, and enzyme, as follows: Column 1, LexA-DHFR, B42, P99 cephalosporinase. Plates in column 1 lack GR and are used as negative controls. Column 2, LexA-DHFR, B42-GR, no enzyme; column 3, LexA-DHFR, B42-GR, P99 cephalosporinase; and column 4, LexA-DHFR, B42-GR, P99 Ser-64 → Ala cephalosporinase. The rows correspond to individual X-Gal plates, which have different small molecules as indicated: No MD, No Mtx-Dex; MD, 1 μM Mtx-Dex; MCD, 10 μM Mtx-Cephem-Dex. (C) ONPG liquid assays. Yeast strains expressing the LexA-DHFR and B42-GR fusion proteins and containing a lacZ reporter gene and expressing no enzyme (left), P99 cephalosporinase (center), or P99 Ser-64 → Ala cephalosporinase (right) were grown in liquid culture and assayed for β-galactosidase activity with ONPG as a substrate. The liquid culture contained small molecules as indicated. The assays were done in triplicate. ONPG hydrolysis rates are reported as nmol/min per mg of total protein, and the error bars for the specific activity correspond to the standard deviation from the mean. Strains containing the active P99 cephalosporinase showed an 8-fold decrease in the level of lacZ transcription relative to strains containing the inactive Ser-64 → Ala variant.