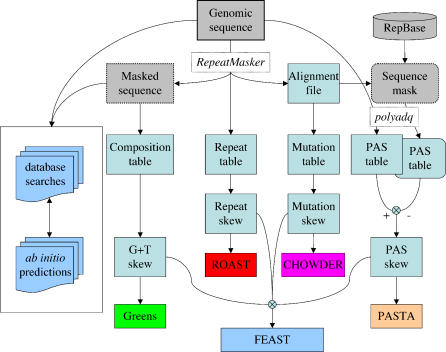
Figure 1. Information Flow in FEAST.
The genomic sequence is analyzed using RepeatMasker, yielding a masked sequence (studied for its base composition), a repeat table, and an alignment file, which is used to list mutations in repeats and to produce a “sequence mask.” Both the original sequence and the sequence mask are studied using polyadq, yielding tables of predicted PASs. The nucleotide composition of the unique sequence, and the mutations within repeats, is tabulated as well. The tables are then analyzed to calculate skews, which are finally used to produce predictive scores, separately for each method (Greens, ROAST, CHOWDER, and PASTA) or in combination (FEAST).