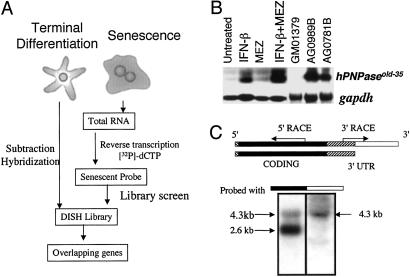
Fig 1.
(A) Schematic representation of the OPS approach. In this application, a differentiation-induction subtraction-hybridization (DISH) library was constructed by subtraction hybridization from terminally differentiated HO-1 human melanoma cells (5). This library was screened with a senescent probe derived from progeria cells. This screening protocol permits the identification of genes displaying parallel (overlapping) changes in expression during terminal differentiation and senescence. (B) Expression of hPNPaseold-35 during terminal differentiation and senescence. Northern blot of hPNPaseold-35 in HO-1 melanoma cells treated for 24 h with IFN-β (2,000 units/ml), MEZ (10 ng/ml) or IFN-β + MEZ (2,000 units/ml + 10 ng/ml), early-passage GM01379 fibroblasts (normal fibroblasts), and two senescent progeroid fibroblasts (AG0781B and AG0989B). gapdh was used as a loading control. (C) Representation of two hPNPaseold-35 transcripts visualized in HO-1 cells on Northern blots. The 5′ UTR (13 bp) in the 5′ region is indicated as a hatched box, the protein-coding region is indicated in black, the 3′ UTR common to both hPNPaseold-35 variants is indicated as a hatched box adjacent to the protein-coding region, and the 3′ UTR present in the longer hPNPaseold-35 variant is indicated in white. The region between the arrows indicates the EST that was identified by using the OPS approach, and the arrows indicate the directions in which the remainder of the gene was cloned. Northern blot analysis of mRNA from HO-1 cells after IFN-β treatment (2,000 units/ml, 7 h) was performed to determine sizes of the hPNPaseold-35 variants. Two membranes containing the same RNA extracts were probed with either the coding region (black, Left) or the 3′ UTR of the longer variant (white, Right).