Abstract
The role of Z-DNA-binding proteins in vivo is explored in yeast. A conformation-specific yeast one-hybrid system is made in which formation of Z-DNA is studied near a minimal promoter site where it can be stabilized by negative supercoiling in addition to protein binding. Experiments were carried out with a Z-DNA-binding protein domain from the editing enzyme, double-stranded RNA adenosine deaminase 1. In the one-hybrid system, the reporter gene is activated when a Z-DNA-specific binding domain is fused with an activation domain and expressed in vivo. Significantly, it was found that even in the absence of the activation domain there is substantial transcription of the reporter gene if the Z-DNA-binding protein is expressed in the cell. This result suggests that Z-DNA formation in the promoter region induced or stabilized by a Z-DNA-binding protein can act as a cis-element in gene regulation. Related results have been found recently when the human chromatin-remodeling system converts a segment of DNA in the promoter region of the human colony-stimulating factor 1 gene into the left-handed Z-conformation.
The yeast one-hybrid system has been used as a tool for identifying proteins that bind to specific DNA sequences (1, 2). The upstream activating sequence of a reporter gene is replaced by a bait sequence, and DNA-binding proteins are then expressed in yeast cells as fusion proteins with a transcriptional activation domain (AD). Thus, transcription occurs only if the DNA-binding protein fused to the AD interacts with the bait DNA sequence. We have expanded this system to include DNA conformational specificity focusing on left-handed Z-DNA formation. This assay allows us to identify and characterize Z-DNA-specific binding proteins in vivo and study the influence of Z-DNA formation on transcriptional activity.
Z-DNA is a left-handed form of the double helix, as revealed in a single-crystal x-ray analysis of duplex d(CGCGCG) (3). The Watson–Crick base pairs in Z-DNA have “flipped over” relative to their orientation in B-DNA, resulting in the guanine residues adopting the syn conformation, whereas the cytosine residues remain in the anti conformation. In addition, it produced a zig-zag arrangement of the sugar phosphate backbone, giving rise to the name Z-DNA. Because purines adopt the syn conformation more readily than pyrimidines, Z-DNA formation is favored in sequences with alternations of purines and pyrimidines. The most favored sequence consists of (dC-dG)n although many other sequences can also adopt the Z conformation (4). Although Z-DNA is a higher energy conformation than right-handed B-DNA, it was realized that B- to Z-DNA conversions could occur in vivo when it was discovered that Z-DNA is stabilized by negative supercoiling (5). Formation of Z-DNA removes negative supercoiling, and the energy of supercoiling stabilizes the Z conformation. Movement of RNA polymerase generates negative torsional strain behind it (6), and Z-DNA is maintained near promoter regions in mammalian nuclei as long as the genes are actively transcribed (7). Furthermore, sequences that form Z-DNA readily occur more frequently near transcription start sites (8). Here we use the yeast one-hybrid system to examine the influence on transcription of Z-DNA formation near the promoter.
Z-DNA can also be stabilized by specific Z-DNA-binding proteins. A protein domain ZαADAR from the editing enzyme double-stranded RNA adenosine deaminase (ADAR1) has been identified and found to bind to Z-DNA with a low nanomolar binding affinity (Kd) (9). The domain has been cocrystallized with Z-DNA (10), and a great deal is known about the ability of this protein and its mutants to bind to Z-DNA (11). If human ZαADAR is added to DNA with a Z-DNA-forming sequence in its center, it will bind to the sequence, even when it is surrounded by B-DNA (12). More recently a domain from the tumor-related protein DLM-1 has also been cocrystallized with Z-DNA and solved (13). This structure is similar to the complex containing human ZαADAR, and it showed that there is a family of such proteins. By sequence analysis, another member of this family is the N-terminal domain of the vaccinia virus E3L protein (9). E3L is an essential virulence factor for the vaccinia virus (14). In this paper we analyze the effect of ZαADAR and several variants in the yeast one-hybrid system. It shows the activity of these proteins in enhancing transcription in a Z-DNA-dependent manner.
In addition, when the AD is eliminated, these proteins bind to Z-DNA-forming sequences and transcription is still activated, although to a lesser extent than is found when the AD is present. These experiments reinforce the observations by Liu et al. (15) who showed that forming Z-DNA near a transcription start site of the human colony stimulating factor-1 (CSF-1) gene favors its transcription.
Materials and Methods
Yeast Strain and Construction of Reporter Plasmids.
Yeast strain YM4271 was purchased from CLONTECH (16) and used for all yeast experiments in this paper. Vector constructs were modified from the vectors used in the MATCHMAKER yeast hybrid system (CLONTECH). Reporter vectors were constructed by using the pLacZi vector as a template and expression vectors were constructed by using either pACT2 or pGAD-GH (CLONTECH).
Target sequences were inserted into the reporter vectors by using duplex DNA oligomers. Oligonucleotides sequences are 5′-CCGAATTCGTCGGT(CG)nACCGACCTCGAGTCTAG AGC-3′ and 5′-GCTCTAGACTCGAGGTCGGT(CG)nACCGACGAATTCGG-3′, where n is 0 for the control. Each pair of oligonucleotides was annealed to form duplex DNA, and it was then digested with appropriate restriction enzymes, EcoRI and XhoI for pLacZi-based vectors. DNAs were purified and ligated to reporter vector DNAs digested with comparable pairs of restriction enzymes.
The LacZ gene reporter constructs pLacZcOp-Control or pLacZcOp-(dC-dG)n (where n is 4, 5, 9, and 12) have a centromere element for self-propagation in yeast. They were made by inserting a PCR-amplified CEN6/ARSH4 containing a DNA fragment from pRS313 between the URA3 gene and the Ampr gene of the pLacZi vector. Each target sequence [control or (dC-dG)n] was inserted into EcoRI and XhoI sites upstream of the LacZ-reporter gene. The resulting pLacZcOp constructs with their target sequence inserts are named pLacZcOp-control and pLacZcOp-(dC-dG)n (Fig. 1). The inserted target sequences were confirmed by dideoxy sequencing. To construct the pLacZcSm vector (Fig. 1), an entire URA3 transcription unit including promoter and terminator was PCR amplified from pLacZcOp with primers, 5′-CGGAATTCAGCACGCCATAGTGACTGG-3′ and 5′-GACTAGCTAGCTCAAGCTTTTCA ATTCATCATTT-3′. The amplified DNA was then cloned into pLacZcOp to replace the existing URA3 gene. The resulting plasmid, pLacZcSm, has the same orientation of transcription in both the URA3 gene and the LacZ gene. The target sequences were then introduced into pLacZcSm with the fragments derived from pLacZcOp-control or (dC-dG)n.
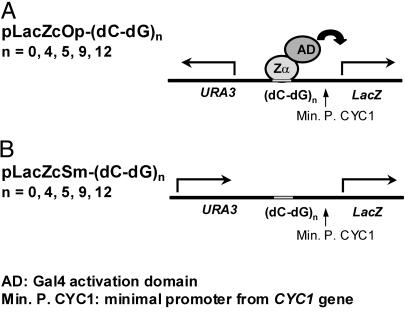
Fig 1.
Schematic representation of reporter vectors. For reporter vector construction, bait sequences of various repeats of (dC-dG) are inserted between the reporter gene (LacZ) and the URA3 gene. Two orientations of the selection marker gene, URA3, are used for creating favorable (A, opposite or Op) or unfavorable (B, same or Sm) conditions for Z-DNA formation. In the pLacZcOp vector, the orientation of URA3 transcription is opposite to that of the LacZ-reporter gene. In contrast, transcriptions from both URA3 and LacZ have the same orientation in the pLacZcSm vector. As shown diagrammatically with pLacZcOp, transcription of the LacZ-reporter gene is activated when hZαADAR carrying an AD binds to Z-DNA. The Zα-AD hybrid fusion is expressed from an independent vector, pACT2-Zα.
Construction of Yeast Expression Vectors for Z-DNA-Binding Proteins With or Without AD Fusion.
Gene fragments of Zα motif proteins were prepared from pET28a (Novagen) bacterial expression constructs. The fragments used were hZαADAR (amino acids 133–209) and hZabADAR (amino acids 133–368) from human ADAR1 (GenBank accession no. U18121), as described elsewhere (17). The engineered Zα motif, hZaaADAR, and the mutant, hZa′bADAR (I172F and N173A in hZabADAR), have been described (12, 18). In addition, hZβADAR (amino acids 294–368) from human ADAR1 was made for this study. NcoI and XhoI digested fragments were inserted into pACT2 at NcoI and XhoI sites. The resulting constructs produce fusion proteins with the Gal4 AD at the N terminus in yeast. The fusion proteins are able to enter the nucleus because the N-terminal simian virus 40 T antigen nuclear localization signal is located at the N terminus of the AD. To construct a plasmid producing proteins without an AD, the Gal4 AD was deleted by KpnI digestion from pGAD-GH (CLONTECH). However, the resulting protein still contains the nuclear localization signal for entering the nucleus. Yeast vector pGNA was made by inserting a linker containing NcoI and XhoI sites at the KpnI site, which allowed the cloning of desired protein genes in yeast.
Yeast One-Hybrid Analysis with Quantitative and Qualitative β-Galactosidase Activity Assay.
Yeast strain YM 4271 was transformed with the LacZ-reporter vectors (either pLacZcOp or pLacZcSm) by using the lithium acetate polyethylene glycol method described (19). The o-nitrophenyl-β-d-galactose (ONPG) assay was used for a quantitative assay of β-galactosidase activity according to the manufacturer's instructions (CLONTECH). Mean and SDs are calculated from triplicates in Miller's β-galactosidase units in which 1 unit hydrolyzes 1 μmol of ONPG per min per cell. For a colorimetric assay of β-galactosidase using plates with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal), freshly transformed yeast cells on selection plates were picked and streaked onto new plates containing 80 μg/ml X-gal and a selection medium lacking uracil and leucine. After incubating at 30°C, a blue color developed if β-galactosidase was produced from the LacZ-reporter gene. pGNA-based vector transformed cells take longer (2–3 d) to develop color than pACT2-based vector cells (1–2 d).
Results
Reporter Vector Constructs Were Designed to Include Z-DNA Stabilized by Negative Supercoiling.
The yeast one-hybrid system was modified to study Z-DNA formation and the role of its binding proteins. Reporter plasmids were constructed containing stretches of alternating dC-dG (Fig. 1). Alternating dC-dG is the sequence with the highest propensity to form Z-DNA, and even short repeats are stable in the Z-conformation in a supercoiled plasmid (20). It has been shown that transcription of RNA from a DNA template produces negative supercoiling behind the moving RNA polymerase (6). The constructs were designed to take advantage of the negative supercoiling produced upstream of an active promoter. The (dC-dG)n sequence was placed between the reporter LacZ gene, with an upstream CYC1 minimal promoter and a constitutively expressed URA3 gene with its promoter in the opposite orientation [Fig. 1A, pLacZcOp-(dC-dG)n (Op)]. The constitutive transcription of URA3 stabilizes (dC-dG)n in the Z-DNA form. As shown diagrammatically in Fig. 1A, binding of the Zα domain of human ADAR1 (hZαADAR) or other Z-DNA-binding proteins fused to the AD should result in the activation of transcription of LacZ. Production of β-galactosidase, the LacZ gene product, can be quantitated by the ONPG assay. The sensitivity of this system is likely to be amplified by expression of the LacZ gene, which will also increase the negative superhelicity of the region between URA3 and LacZ, thereby enhancing Z-DNA formation.
An additional plasmid, pLacZcSm-(dC-dG)n (Sm) shown in Fig. 1B, was constructed as a control to assess the Z-DNA specificity of binding. Positive supercoiling generated from transcription of URA3 gene by RNA polymerase produces an unfavorable environment for Z-DNA formation at the (dC-dG)n sequence. Proteins specific for the B-DNA conformation of (dC-dG)n sequence will bind Sm as well as or better than Op, depending on whether Z-DNA formation is inhibitory to their sequence-specific binding. In contrast, Z-DNA-binding proteins will bind preferentially in the Op orientation, with binding to Sm dependent on the ability of the Z-DNA-binding protein to independently stabilize Z-DNA.
The length of the alternating dC-dG sequence can affect the results of these experiments. There are optimal sizes for both Z-DNA formation and protein binding. The Op plasmids were transfected into yeast (i) alone, (ii) with a plasmid expressing only an AD (pACT2), or (iii) with a plasmid expressing the AD attached to hZαADAR. Fig. 2A shows that the amount of β-galactosidase activity increases approximately linearly with increasing number of dC-dG repeats in the presence of Zα attached to an AD. In the absence of hZαADAR, either with or without pACT2, there is only minimal enzyme activity if the number of repeats is less than five. Interestingly, at higher repeat numbers there is a slight increase in activity, even in the absence of an AD. This suggests that formation of Z-DNA near a promoter may in itself act as an enhancer. This observation is investigated further and discussed below.
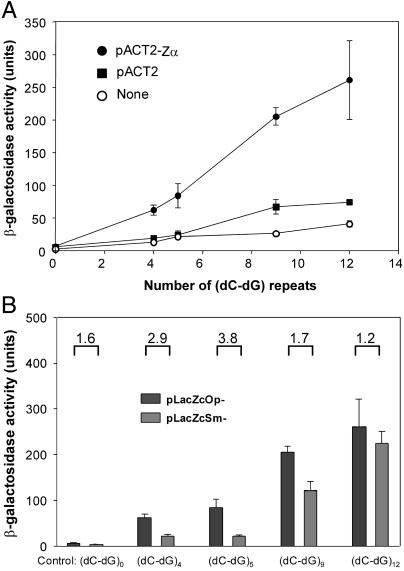
Fig 2.
The Zα-AD fusion protein activates transcription of the LacZ-reporter gene by binding to upstream Z-DNA forming bait sequences. (A) Units of β-galactosidase activity made as a function of n, the number of (dC-dG) repeats in the vector; n = 0 represents a control sequence. pLacZcOp vectors were transfected into yeast either without (None) or with pACT2 or the pACT2-Zα expression vectors containing hZαADAR. β-galactosidase activity measured by the quantitative ONPG assay shows that an increasing number of repeats of (dC-dG) in the bait sequences give rise to an increase of reporter gene activation. Three measurements were made at each point. (B) Comparison of activation of the LacZ gene by the Zα-AD fusion protein in different reporter vectors, pLacZcOp and pLacZcSm. The numbers above the bars represent the fold increase of the enzyme activity in pLacZcOp compared to pLacZcSm. The differences are significant with (dC-dG)4 and (dC-dG)5 but not with (dC-dG)9 and (dC-dG)12.
β-galactosidase is induced in the presence of hZαADAR in the Sm plasmid as well as Op (Fig. 2B). The induction in Sm is likely to reflect the fact that the high-affinity hZαADAR is sufficient to stabilize the Z-conformation in dC-dG repeats even within a B-DNA environment (12). Further, comparison of induction of activity in Op and Sm shows an interesting relationship. Although induction is roughly comparable at high repeat number, there is a significant difference at shorter lengths. Op shows dramatic induction in (dC-dG)4 containing plasmids, whereas Sm has only modest enzyme activity below (dC-dG)9. The plasmids containing (dC-dG)4 and (dC-dG)5 contain only a single hZαADAR-binding site per strand, based on the crystal structure of the hZαADAR–Z-DNA complex (10). This may not be enough to stabilize Z-DNA in the absence of supercoiling. The longer repeats contain multiple binding sites, which can act together to stabilize Z-DNA even in the absence of supercoiling. The same phenomenon was demonstrated in vitro; the cleavage by a Z-DNA-specific nuclease was enhanced in a sequence containing multiple Zα-binding sites, in the presence of excess hZαADAR (12).
hZβADAR Does not Bind Z-DNA.
Although hZαADAR is a stable protein domain by itself, partial proteolysis of the N terminus of human ADAR1 shows that it is included in a larger domain, called hZabADAR (17). This domain consists of hZαADAR, as well as a second region, hZβADAR, closely related by sequence. In addition there is an intervening linker, which is tandemly duplicated in humans. The in vitro binding of Z-DNA by hZabADAR resembles but is not identical to that of hZαADAR (17). Although hZβADAR is very similar in sequence to hZαADAR, it does not bind Z-DNA in vitro (Y.-G.K., unpublished data).
A number of constructs have been designed to examine the binding of hZabADAR to DNA and the roles of the two related subdomains. In hZa′bADAR, a double mutation in hZαADAR (I172F and N173A) virtually abolishes its ability to bind Z-DNA in vitro (12). hZaaADAR contains a second hZαADAR motif replacing hZβADAR in hZabADAR (18). hZaaADAR binds Z-DNA with a higher affinity than either hZαADAR or hZabADAR.
Examination of in vivo binding of Z-DNA by these various peptides agrees well with the in vitro results (Fig. 3). Fig. 3A shows activation of the LacZ gene by different peptides in the presence of varying lengths of alternating (dC-dG). hZβADAR and hZa′bADAR show no significant increase of activation above the pACT2 control, whereas hZαADAR, hZabADAR, and hZaaADAR all produce activation of the LacZ gene. When the reporter construct contains only four or five repeats of (dC-dG), all three of these proteins behave identically. With longer repeats, hZabADAR activates LacZ in a manner similar to hZαADAR, whereas hZaaADAR is dramatically better. In hZaaADAR, it is likely that the two copies of hZαADAR act cooperatively when positioned next to each other to produce a higher level of activation.
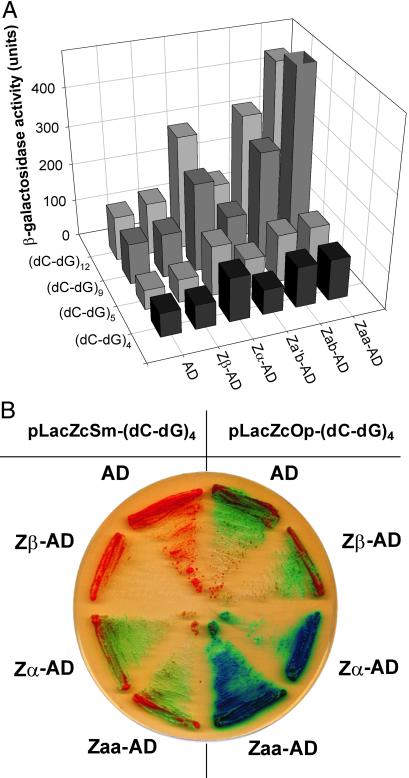
Fig 3.
Analysis of the Z-DNA-binding activities of Zα, Zβ, Za′b, Zab, and Zaa by using lacZ-reporter gene activation. Zα, Zβ, and Zab all come from human ADAR1 (17). In Zaa, the Zβ domain of Zab is removed and Zα replaces it (18). Za′b has mutations in Zα of Zab as described (12). (A) Vectors containing an AD fused to nothing (pACT2) or to various human Zα constructs from human ADAR1 (pACT2-Zβ, -Zα, -Zab, -Za′b, and –Zaa) were cotransfected into yeast with pLacZcOp- (dC-dG)n (where n = 4, 5, 9, and 12). β-galactosidase activities were determined by quantitative ONPG assay. In general, background enzyme activity (with pACT2 transfection) increases as longer repeats of (dC-dG) are used in bait sequences. The hZαADAR-containing peptides (hZαADAR, hZabADAR, and hZaaADAR) show high levels of enzyme activity. However, hZβADAR and hZa′bADAR do not show significant activity above the controls. (B) Yeast cells were transformed with recombinant pACT2-based vectors and either pLacZcOp-(dC-dG)4 or pLacZcSm-(dC-dG)4 and plated onto selective medium containing X-gal, a β-galactosidase substrate. Development of a blue color indicates β-galactosidase activity hydrolyzing X-gal, as described in Materials and Methods. Higher levels of β-galactosidase activity were shown in pLacZcOp-(dC-dG)4 than in pLacZcSm-(dC-dG)4.
When reporter vectors Op and Sm containing four repeats of dC-dG are compared on X-gal plates, it is clear that the presence of a Z-DNA-binding domain, either hZαADAR or hZaaADAR, with divergent promoters (Op) produce the most β-galactosidase activity (Fig. 3B). Even in the absence of stabilizing negative superhelicity (Sm), hZαADAR and hZabADAR have some stimulatory effect, as was previously shown in Fig. 2B. The presence of hZβADAR has no effect, indicating that hZβADAR does not bind to the promoter region. The level of β-galactosidase activity in the presence of hZβADAR is the same as the AD alone. This clearly indicates that hZβADAR does not bind Z-DNA in vivo in agreement with in vitro results (Y.-G.K., unpublished data).
Z-DNA Formation Stabilized by Zα near a Promoter Region Acts as an Enhancer Element.
Z-DNA-forming sequences are common near transcription start sites (8). Liu et al. (15) have suggested that Z-DNA, occurring near a promoter, might have a regulatory effect on that promoter. To detect such an effect, we have used hZαADAR as a tool to stimulate the formation of Z-DNA in vivo. hZαADAR without an AD was expressed in yeast cells containing Op- or Sm-reporter constructs. In the absence of an AD, hZαADAR cannot activate β-galactosidase expression by interacting with the promoter, unless the presence of Z-DNA per se has an effect on transcription of a nearby promoter. Comparison of results from different reporter constructs expressing either hZαADAR or hZaaADAR shows that increasing Z-DNA formation is proportional to reporter gene expression (Fig. 4). In the absence of hZαADAR expression, longer repeats of (dC-dG) yield slightly greater β-galactosidase expression, as noted in Fig. 2A. This increased level of transcription in the presence of longer repeats of (dC-dG) is greatly amplified by the expression of hZαADAR that is known to facilitate Z-DNA formation (Fig. 4A). This effect is even more dramatic in an environment unfavorable for Z-DNA formation (Fig. 4B). With (dC-dG)5, activation of the LacZ-reporter gene is shown only if Z-DNA formation is induced by hZαADAR or hZaaADAR in the Op template. However, a longer stretch of (dC-dG) provides more binding sites for hZαADAR so that Z-DNA is formed even in the presence of positive supercoiling produced from the URA3 promoter of the Sm plasmid (Fig. 4C). Thus, induction of Z-DNA by hZαADAR can activate reporter gene transcription. Similar effects are seen for hZabADAR (data not shown).
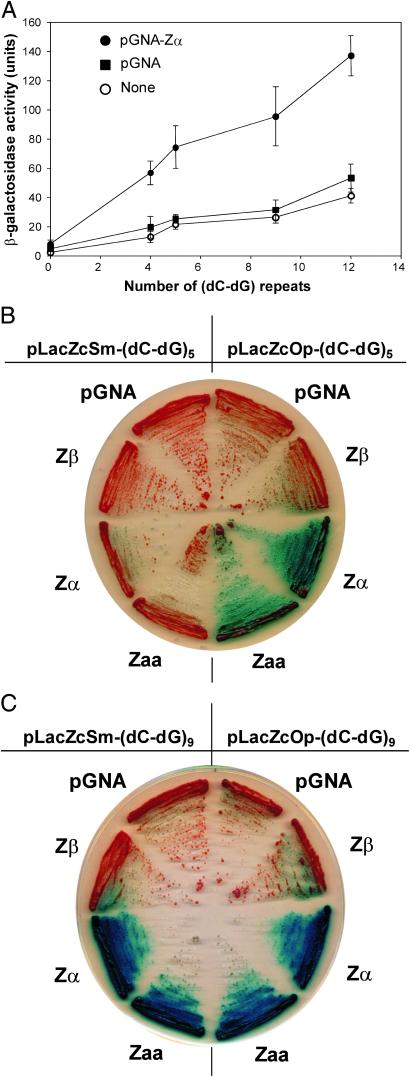
Fig 4.
Zα by itself activates transcription of the reporter. Expression of Zα alone without the Gal4 AD was examined. pGNA-Zα expresses Zα with the simian virus 40 T-antigen nuclear localization signal at the N terminus (described in Materials and Methods). (A) Experiments were performed with different pLacZcOp-(dC-dG)n reporters. hZαADAR expression increases β-galactosidase activity significantly as stretches of dC-dG become longer. Activation on X-gal plates of the reporter gene LacZ by the pGNA plasmid alone or expressing various proteins; pGNA-Zβ, pGNA-Zα, and pGNA-Zaa in pLacZcOp-(dC-dG)n or pLacZcSm-(dC-dG)n when n = 5 (B) or n = 9 (C). The contribution of negative supercoiling to Z-DNA formation because of opposite or same orientations is apparent in B, where n = 5. The great tendency of longer stretches to form Z-DNA where n = 9 swamps out the smaller distinction between opposite and same orientation.
Discussion
The hZαADAR and hZabADAR domains of human ADAR1 have been extensively characterized and much is known about their affinity for Z-DNA under a variety of in vitro conditions (9, 12, 17, 18, 21, 22). Nevertheless, conditions existing in the eukaryotic nucleus are quite different from those in a test tube. The results reported here clearly demonstrate that binding to Z-DNA by hZαADAR, as well as its homologs and various engineered constructs, closely resemble the conformational specificity in vitro. Evidence for the Z-DNA specificity in vivo of both hΖαADAR and hZabADAR is provided by the enhanced expression of β-galactosidase when ura3 and lacZ are organized in opposite directions (Fig. 3). A protein binding to B-DNA would not produce these results. The in vivo assay provides extra information because it is more nuanced than routine in vitro assays. In vivo, a semiquantitative hierarchy of binding affinity of hZαADAR and its homologs can be seen, which is not apparent from in vitro CD and the electrophoretic mobility-shift assays.
Limited proteolysis of the N terminus of ADAR1 showed that hZabADAR is a compact, stable domain (17). hZabADAR binds Z-DNA with a Kd equivalent to that of hZαADAR, and there is no other Z-DNA-binding region in hZabADAR (17). It is possible that the linker and hZβADAR regions are irrelevant to the hZabADAR–Z-DNA interaction. On the other hand, hZabADAR has more sequence specificity than hZαADAR, suggesting that hZβADAR may make some DNA contact that changes sequence preference (17, 18). However, the function of hZβADAR is unknown; strikingly, in zebra fish ADAR1 the homologous region contains two Zα-like domains each capable of binding Z-DNA (Y.-G.K., unpublished data).
Liu et al. (15) showed that during activation of the CSF1 promoter Z-DNA formation is induced by the chromatin-remodeling system, the mammalian SWI/SNF or BAF complex. It is known that chromatin-remodeling systems release negative superhelicity (23, 24), which can provide the energy for Z-DNA formation. Because Z-DNA cannot go into nucleosomes (25), Liu et al. (15) suggested that Z-DNA formation thus keeps chromatin structure open, facilitating the assembly of transcriptional machinery. This model suggests an explanation of why there is some enhancement of β-galactosidase expression in the Op construct, even in the absence of a Z-DNA-binding protein (Fig. 2A).
This hypothesis has additional implications if one considers that hZαADAR is able to induce formation of Z-DNA in a suitable sequence when it is surrounded by B-DNA both in vitro (12) and in vivo as shown here. When hZαADAR is expressed alone in this assay system, it is sufficient to activate transcription of the reporter gene in a Z-DNA-dependent manner. In Fig. 4A, when there are no (dC-dG) repeats, expressing hZαADAR has no effect. However, in the presence of larger numbers of (dC-dG) repeats, there is some activation of the reporter gene even without hZαADAR expression. But with expression of hZαADAR, the level of reporter gene activation increases significantly. It is possible that hZαADAR, by binding to the (dC-dG) repeats and inducing Z-DNA, may help maintain an open chromatin structure in the surrounding promoter area leading to the assembly of a transcription initiation complex and increased transcription. Furthermore, the Z-DNA conformation will be maintained while transcription is continuing as negative torsional strain continues to be fed into the region. The segment reverts to B-DNA only when transcription is down-regulated (7). An increase in basal level activity of the reporter gene is observed even without hZαADAR expression, especially when Z-DNA formation is favored in the bait sequences (Fig. 2A) either by longer (dC-dG) repeats or the accumulation of negative supercoiling resulting from transcription of two genes with promoters in opposite directions. This phenomenon is amplified by further induction of Z-DNA formation by hZαADAR (Fig. 4A). As with the CSF1 gene (15), Z-DNA may thus be one of the factors that govern the availability of target sites, especially in the initial steps in transcription. The likelihood of Z-DNA formation usually depends on its environment, particularly negative supercoiling derived from many cellular activities. Recent data from studies of chromatin remodeling complexes adds another mechanism for generating negative supercoiling (23). Z-DNA formation can thus be a cis-element-influencing chromatin structure.
Finally, it is possible that the formation of Z-DNA and resulting changes in chromatin structure could affect activities other than transcription, such as recombination or DNA repair. Identification of novel proteins that interact with Z-DNA may thus gain a new perspective. Understanding their binding specificities may help us to understand their modes of operation. The conformation-specific yeast one-hybrid system is a valuable tool in the search for such proteins.
Acknowledgments
We thank Ky Lowenhaupt for helpful discussion. This research was supported by grants from the National Science Foundation, the National Institutes of Health, and the National Foundation for Cancer Research.
Abbreviations
ADAR1, double-stranded RNA adenosine deaminase 1
Op, pLacZcOp
Sm, pLacZcSm
AD, activation domain
ONPG, o-nitrophenyl-β-d-galactose
X-gal, 5-bromo-4-chloro-3-indolyl-β-d-galactoside
References
- 1.Liu J., Wilson, T. E., Milbrandt, J. & Johnston, M. (1993) Methods 5, 125-137. [Google Scholar]
- 2.Allen J. B., Walberg, M. W., Edwards, M. C. & Elledge, S. J. (1995) Trends Biochem. Sci. 20, 511-516. [DOI] [PubMed] [Google Scholar]
- 3.Wang A. H., Quigley, G. J., Kolpak, F. J., Crawford, J. L., van Boom, J. H., van der Marel, G. & Rich, A. (1979) Nature 282, 680-686. [DOI] [PubMed] [Google Scholar]
- 4.Rich A., Nordheim, A. & Wang, A. H. (1984) Annu. Rev. Biochem. 53, 791-846. [DOI] [PubMed] [Google Scholar]
- 5.Peck L. J., Nordheim, A., Rich, A. & Wang, J. C. (1982) Proc. Natl. Acad. Sci. USA 79, 4560-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L. F. & Wang, J. C. (1987) Proc. Natl. Acad. Sci. USA 84, 7024-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittig B., Wolfl, S., Dorbic, T., Vahrson, W. & Rich, A. (1992) EMBO J. 11, 4653-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroth G. P., Chou, P. J. & Ho, P. S. (1992) J. Biol. Chem. 267, 11846-11855. [PubMed] [Google Scholar]
- 9.Herbert A., Alfken, J., Kim, Y. G., Mian, I. S., Nishikura, K. & Rich, A. (1997) Proc. Natl. Acad. Sci. USA 94, 8421-8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz T., Rould, M. A., Lowenhaupt, K., Herbert, A. & Rich, A. (1999) Science 284, 1841-1845. [DOI] [PubMed] [Google Scholar]
- 11.Schade M., Turner, C. J., Lowenhaupt, K., Rich, A. & Herbert, A. (1999) EMBO J. 18, 470-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y. G., Lowenhaupt, K., Maas, S., Herbert, A., Schwartz, T. & Rich, A. (2000) J. Biol. Chem. 275, 26828-26833. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz T., Behlke, J., Lowenhaupt, K., Heinemann, U. & Rich, A. (2001) Nat. Struct. Biol. 8, 761-765. [DOI] [PubMed] [Google Scholar]
- 14.Brandt T. A. & Jacobs, B. L. (2001) J. Virol. 75, 850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu R., Liu, H., Chen, X., Kirby, M., Brown, P. O. & Zhao, K. (2001) Cell 106, 309-318. [DOI] [PubMed] [Google Scholar]
- 16.Wilson T. E., Fahrner, T. J., Johnston, M. & Milbrandt, J. (1991) Science 252, 1296-1300. [DOI] [PubMed] [Google Scholar]
- 17.Schwartz T., Lowenhaupt, K., Kim, Y. G., Li, L., Brown, B. A., II, Herbert, A. & Rich, A. (1999) J. Biol. Chem. 274, 2899-2906. [DOI] [PubMed] [Google Scholar]
- 18.Kim Y. G., Lowenhaupt, K., Schwartz, T. & Rich, A. (1999) J. Biol. Chem. 274, 19081-19086. [DOI] [PubMed] [Google Scholar]
- 19.Gietz R. D., Schiestl, R. H., Willems, A. R. & Woods, R. A. (1995) Yeast 11, 355-360. [DOI] [PubMed] [Google Scholar]
- 20.Kim J., Yang, C. & DasSarma, S. (1996) J. Biol. Chem. 271, 9340-9346. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y. G., Kim, P. S., Herbert, A. & Rich, A. (1997) Proc. Natl. Acad. Sci. USA 94, 12875-12879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown B. A. & Rich, A. (2001) Acta Biochim. Pol. 48, 295-312. [PubMed] [Google Scholar]
- 23.Havas K., Flaus, A., Phelan, M., Kingston, R., Wade, P. A., Lilley, D. M. & Owen-Hughes, T. (2000) Cell 103, 1133-1142. [DOI] [PubMed] [Google Scholar]
- 24.Gavin I., Horn, P. J. & Peterson, C. L. (2001) Mol. Cell 7, 97-104. [DOI] [PubMed] [Google Scholar]
- 25.Garner M. M. & Felsenfeld, G. (1987) J. Mol. Biol. 196, 581-590. [DOI] [PubMed] [Google Scholar]