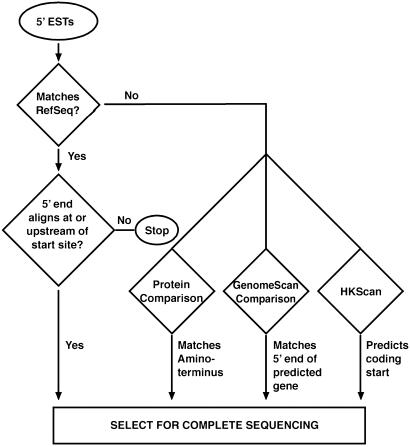
Fig. 1.
Tests for identifying putative full-ORF cDNA clones. In the first test, 5′ ESTs first were compared with all available ORF-complete mRNA sequences from the same organism (human or mouse) in the RefSeq collection. When a 5′ EST aligned (>95% homology for 100 or more base pairs) at or upstream of an annotated translation start site, that clone was considered to contain a candidate full-ORF cDNA. However, if the 5′ EST aligned downstream from an annotated translational start site, that clone was eliminated from consideration, although some of these may be full-ORF clones with an alternate 5′ translational start site. Any 5′ ESTs that did not match a RefSeq sequence were subjected to additional tests. In the second test, six possible frame translations were compared with the subset of GenBank protein records originating from Protein Information Resource (15), Protein Data Base (16), or SwissProt (17) that begin with methionine. This test identifies ESTs from genes with an N terminus similar but not identical to a known protein. Thus, in cases where a protein match (<90% identity but with an E value of less than or equal to 10−6) was detected and incorporated the known initiating methionine, the associated cDNA clone was considered a candidate to have a complete ORF. In the third test, we compared each 5′ EST to a collection of predicted genes derived from the human genome sequence by genomescan (18). When a 5′ EST aligned (95% identity for 100 or more bp) to a gene prediction that begins with ATG, the associated clone was considered a candidate. In the fourth test, we used the new program hkscan, which looks for evidence of a transition from noncoding to coding sequence (described in Materials and Methods).