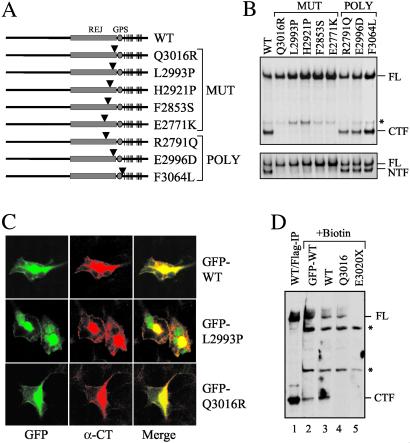
Fig 3.
Effect of PKD1-associated mutations on cleavage. (A) Schematic structures of five disease-associated mutant forms of polycystin-1 and three normal variants. An arrowhead marks the relative position of the substitution in each protein. “MUT” identifies disease-associated mutations and “POLY” identifies polymorphic variants. (B) The cleavage property of each mutation was analyzed by using either α-CT (Upper) or α-LRR (Lower). The asterisk indicates an additional band of uncertain nature. (C) Cleavage-deficient forms of polycystin-1 traffic normally. GFP-WT, GFP-L2993P, and GFP-Q3016R were expressed in HEK cells and evaluated for the subcellular localization of each. (D) A cell surface biotinylation study was performed on HEK cells transfected with various constructs as indicated. Biotinylated surface proteins were captured by streptavidin beads and analyzed on Western blot using α-CT. WT/Flag-IP without biotin treatment (lane 1) served as control for FL and CTF. E3020X served as negative control for α-CT. Asterisks indicate nonspecific bands of unknown identity.