Abstract
Real-time PCR has been developed to genotype measles virus (MV) isolates. MV strains circulating in epidemics in Gabon in 1984, Cameroon in 2001, Morocco in 2003, and France in 2004 were investigated. We developed a real-time amplification refractory mutation system PCR (RT-AMRS PCR) using SYBR green fluorescent dye. Six pairs of primers for RT-ARMS PCR were designed to specifically amplify genotypes A, B2, B3.1, B3.2, C2, and D7. Genotypes could be differentiated by melting curve analysis. All strains were also confirmed by direct sequencing. Using the result obtained by direct sequencing and phylogenetic analysis as the reference, the accuracy of MV by RT-ARMS PCR and melting curve analysis was 97%. However, the latter method is more rapid and sensitive than the former method. This method could be a useful tool for molecular epidemiological studies of MV, providing an efficient alternative for large-scale studies.
Measles virus (MV) infection is a public health problem worldwide, with an estimated 30 million people infected and 530,000 deaths in 2003 (21). MV is serologically a monotypic virus; however, diversity within the complete hemagglutinin (H) gene and the C-terminal part of the nucleoprotein (N) gene has allowed the classification of MV into eight clades, designated A to H, including 22 genotypes (22). Measles has been designated as a target for eradication by the WHO by the year 2010. According to a strategic plan for the elimination of measles, molecular epidemiology will play an essential role in measuring the impact of the vaccination campaign in countries where the disease remains endemic and in tracing the source of MVs isolated in countries with an advanced eradication program, where most cases are imported.
The WHO recommends the genotyping of representative strains in every outbreak. This is essentially performed by nucleotidic sequencing of genomic fragments followed by phylogenetic analysis. Although this method is the “gold standard” and allows direct classification, it is time-consuming and expensive and does not allow rapid analysis of numerous samples. Alternative methods have been described, such as restriction fragment length polymorphism and nucleotide-specific multiplex PCR to differentiate MV strains (11, 18). These methods rely on gel electrophoreses analysis that can lead to ambiguous interpretations. Samuel et al. (17) developed a method based on a modification of the amplification refractory mutation system, which was limited to a single genotype and needed a post-PCR enzyme-linked immunosorbent assay analysis, which is time-consuming.
In this study, a method for genotyping measles virus was performed by real-time amplification refractory mutation system (RT-ARMS) PCR using SYBR green fluorescent dye. The method uses genotype-specific primers to specifically amplify MV genotypes. This novel assay allows real-time detection of PCR products, and the genotype is subsequently determined by melting curve analysis. In the present studies, RT-ARMS PCR was used to genotype 30 isolates from MV outbreaks in Africa and France. The results using this method were confirmed by comparison to the direct sequencing technique with the same sample. The data show that this new method has the advantages of rapidity, reproducibility, and accuracy, which would be both feasible and attractive for large-scale use.
MATERIALS AND METHODS
Patients and virus isolation.
Viruses were isolated from clinical specimens of patients with clinically diagnosed measles during different epidemics. Three clinical specimens were from Libreville, Gabon, in 1984, and 17 specimens were from Yaounde, Cameroon, in 2001. The sequences of certain of these strains have been previously published (9, 19). Seven urine samples were collected at the time of measles outbreaks in Casablanca, Taroudant, and other locations within Morocco in 2003. In addition, three samples were collected from sporadic cases during 2004 in France.
One sample was isolated in Caen (Caen-04) from an adult, and the two others were isolated in Lyon (Lys04-1 and Lys04-2). Strain Lys04-1 was isolated from a patient after a vacation in Africa, and strain Lys04-2 was isolated from a child after measles-mumps-rubella vaccination.
Samples were taken within 6 days after the onset of the rash. Peripheral blood mononuclear cells were isolated by Ficoll gradient centrifugation and cocultivated either with Vero cells for the Gabon strains from 1984 or with B95a cells for other samples in Dulbecco's modified Eagle's medium supplemented with 2% fetal calf serum. Viruses were stored at −80°C.
Further MV strains were used to set up the RT-ARMS PCR assay. They are summarized in Tables 1 and 2.
TABLE 1.
MV strains used to establish the RT-ARMS PCR
| Strain | Description | Genotype | Reference or source |
|---|---|---|---|
| Edmonston | Vaccine strain | A | 10 |
| Halle | Vaccine strain | A | 4 |
| Rouvax | Vaccine strain | A | 6 |
| Zagreb | Vaccine strain | A | 14 |
| R113 | MVi/Libreville.GAB/84 (R113) | B2 | 19 |
| R115 | MVi/Libreville.GAB/84 (R115) | B2 | This study |
| Ibadan 10 | MVi/Ibadan.NIE/10.98 | B3.1 | 9 |
| CR81 | MVi/Yaounde.CAM/8.01/6 | B3.1 | 19 |
| Lys | MVi/Lyon.FRA/94 | B3.2 | 7 |
| G954 | MVi/Gambia/93 | B3.2 | 9 |
| M48 | MVi/Temara.MOR/6.03 | C2 | This study |
| M185 | MVi/Sefrou.MOR/24.03/1 | C2 | This study |
| Lys03 | MVi/Lyon.FRA/03 | D7 | 23 |
| Pa01 | MVi/Paris.FRA/01 | D7 | 23 |
TABLE 2.
Genotypes and GenBank accession numbers of the different strains analyzed in this study
| Strain | Description | Genotype | Accession numbera
|
|
|---|---|---|---|---|
| N gene | H gene | |||
| CR63 | MVi.Yaounde.CAE/6-1.01 | B3.1 | DQ267506 | ND |
| CR64 | MVi.Yaounde.CAE/6-2.01 | B3.1 | DQ267507 | ND |
| CR65 | MVi.Yaounde.CAE/6-3.01 | B3.1 | DQ267508 | ND |
| CR66 | MVi.Yaounde.CAE/6-4.01 | B3.1 | DQ267509 | ND |
| CR68 | MVi.Yaounde.CAE/6-6.01 | B3.1 | DQ267510 | ND |
| CR69 | MVi.Yaounde.CAE/6-7.01 | B3.1 | DQ267511 | ND |
| CR70 | MVi.Yaounde.CAE/6-8.01 | B3.1 | DQ267512 | ND |
| CR76 | MVi.Yaounde.CAE/8-1.01 | B3.1 | DQ267513 | ND |
| CR77 | MVi.Yaounde.CAE/8-2.01 | B3.1 | DQ267514 | ND |
| CR78 | MVi.Yaounde.CAE/8-3.01 | B3.1 | DQ267515 | ND |
| CR79 | MVi.Yaounde.CAE/8-4.01 | B3.1 | DQ267516 | ND |
| CR80 | MVi.Yaounde.CAE/8-5.01 | B3.1 | DQ267517 | ND |
| CR81 | MVi.Yaounde.CAE/8-6.01 | B3.1 | DQ267518 | ND |
| Lys04-1 | MVi.Lyon.FRA/20.04/1 | B3.1 | DQ267519 | DQ267504 |
| Lys04-2 | nvs.Lyon.FRA/20.04/2 | A | DQ267520 | ND |
| Caen04 | MVi.Caen.FRA/04 | B3.1 | DQ267521 | DQ267505 |
| R102 | MVi.Libreville.GAB/84(R102) | B2 | DQ267522 | ND |
| R112 | MVi.Libreville.GAB/84(R112) | B2 | DQ267523 | ND |
| R115 | MVi.Libreville.GAB/84(R115) | B2 | DQ267524 | ND |
| M48 | MVi/Sefrou.MOR/6.03 | C2 | DQ267525 | DQ267497 |
| M76 | MVi/Kelâa.MOR/13.03 | C2 | DQ267526 | DQ267498 |
| M140 | MVi/Casablanca.MOR/19.03/1 | C2 | DQ267527 | DQ267499 |
| M141 | MVi/Casablanca.MOR/19.03/2 | C2 | DQ267528 | DQ267500 |
| M148 | MVi/Taroudant.MOR/19.03/3 | C2 | DQ267529 | DQ267501 |
| M150 | MVi/Taroudant.MOR/19.03/4 | C2 | DQ267530 | DQ267502 |
| M185 | MVi/Temara.MOR/24.03/1 | C2 | DQ267531 | DQ267503 |
ND, not done.
Measles virus genotyping by sequence analysis.
All samples analyzed by RT-ARMS PCR were initially genotyped by sequence analysis. Viral RNA was extracted from the infected cell supernatant using the RNA extraction kit (QIAamp Viral RNA Mini kit; QIAGEN Inc., Valencia, CA) according to the manufacturer's instructions. The reverse transcriptase PCR for the N and H genes was carried out, and the amplicons were purified and sequenced as previously described (9). Multiple alignment of nucleotide sequences and construction of phylogenetic trees were carried out using the Clustal X program, version 1.81 (8), and confirmed with 1,000 bootstrap replicates.
Design of genotype-specific primer for RT-ARMS PCR.
To identify specific single-nucleotide polymorphisms (SNPs) (also called genotype-specific nucleotides), for genotype differentiation, we studied the multiple alignments of 450 nucleotides of the C-terminal region of nucleoprotein gene sequences, which are currently available from the viral reference genome database (http://www.ncbi.gov). For each genotype identified, one or more genotype-specific nucleotides were identified. A pair of primers, each with an SNP at the 3′ end for genotypes A, B2, B3.1, and C2, was designed. For genotypes B3.2 and D7, only one primer contains the SNP, as we were unable to find an appropriate SNP for the second primer. To optimize the amplification during the RT-ARMS PCR, the primers were chosen to yield a product between 100 and 200 bp. Based on the principle described previously by Newton et al. (13) and modified by Bai and Wong (2), to increase the specificity of the RT-ARMS reaction, mismatches were introduced at the two nucleotides immediately 5′ to the SNP. Hence, during the annealing process, the primers would match with targets containing perfectly complementary sequence rather than with the ones containing the mismatched signature SNP.
N-gene first-round amplification.
Viral RNA was extracted from infected cell supernatants using the QIAamp Viral RNA Mini kit (QIAGEN). Specific cDNA of the N gene was synthesized by reverse transcription at 55°C for 30 min, immediately followed by PCR amplification in the same tube, using SuperScript One-Step reverse transcription-PCR with platinum Taq (Invitrogen) according to the manufacturer's instructions.
For the N gene, primers MVNPCR2 (nucleotides 975 to 996 [5′-GCTGGTGAGTTATCCACACTTG-3′]) and MVNPCR4 (nucleotides 1701 to 1722 [5′-GTAGGCGGATGTTGTTCTGGTC-3′]) were used to amplify a 747-bp fragment. The PCR cycling program consisted of a denaturation step for 4 min at 94°C followed by 35 cycles of 30 s at 94°C, 45 s at 55°C, and 45 s at 72°C with a final extension step for 7 min at 72°C. All products were held at 4°C. These products were used as a target for RT-ARMS PCR.
Genotyping of MV by RT-ARMS PCR.
Previously genotyped samples served as controls for developing the genotyping assay based on RT-ARMS PCR and melting curve analysis. RT-ARMS PCR was performed using the ABI PRISM 7000 sequence detection system (Applied Biosystems). A master mix reaction mixture was prepared and dispensed in 15-μl aliquots into thin-walled microAmp optical tubes (ABI PRISM; Applied Biosystems). Five microliters of N-gene PCR product was then added to each tube. The final reaction mixture contained 400 nM of each primer, 12.5 μl of 2× Platinum SYBR green qPCR SuperMix initial uracil DNA glycosylase, 0.5 μl Rox reference dye (Invitrogen), and RNase-free water to complete the reaction mixture volume to 20 μl. All reactions were performed in duplicate. To test the sensibility of the RT-ARMS PCR, first reactions were assayed using three different dilutions of target containing 0.26 ng, 2.60 ng, or 26 ng of PCR product.
The PCR was performed with an initial uracil DNA glycosylase decontamination step at 50°C for 2 min followed by a hot-start denaturation step at 95°C for 10 min. Next, PCR amplification was carried out for 40 cycles at 95°C for 15 s and 60°C for 1 min. The fluorescence was read during the reaction, allowing a continuous monitoring of the amount of PCR product. After amplification, melting curve analysis was performed on the product by heating from 60°C to 95°C for 20 min.
The real-time PCR data were analyzed with ABI PRISM 7000 SDS v1.1 data analysis software (Applied Biosystems). The melting temperature (Tm) indicated by the derivative melting curves in each sample was used to identify the MV genotype. The samples whose melting curves shared the same Tm point with the control genotypes were interpreted as belonging to the same genotype.
Nucleotide sequence accession numbers.
Nucleotide sequences for MV strains analyzed in this study have been deposited in the GenBank database under the accession numbers listed in Table 2.
RESULTS
MV genotyping by sequence analyses.
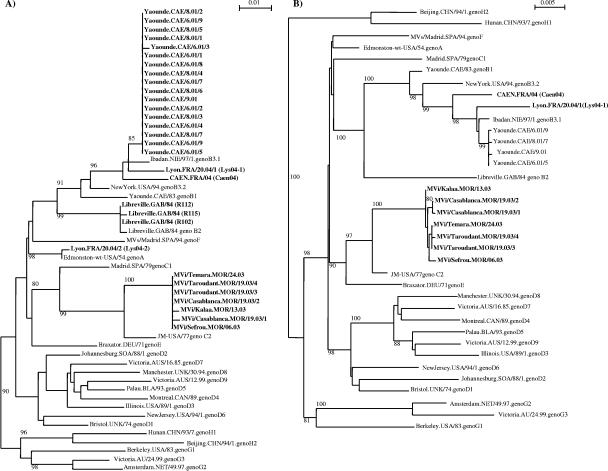
MV RNA from isolates was amplified (30 isolates for the N protein and 9 isolates for the H protein) by PCR. The sequence analyses of these isolates (Fig. 1) showed that the Gabon measles strains were genotype B2, as previously described (19). All Cameroonian strains were highly related to each other. At the nucleotide level, there was less than 0.2% heterogeneity within the N gene. Phylogenetic analyses showed that they belonged to genotype B3.1. These studies confirmed our previous results in which we sequenced four strains (Hand N genes) from the same epidemic (9).
FIG. 1.
Phylogenetic analysis of measles isolates based on the N gene (A) and the H gene (B). Significant bootstrap values (>80%) are indicated.
All samples collected during outbreaks in Morocco were related to each other and to the samples circulating during the 1998 and 1999 epidemics (1), as the nucleotide heterogeneity was less than 0.2% for the N gene and 0.5% for the H gene. They were all determined to be genotype C2. Since the initial studies of molecular characterization of measles viruses in Morocco in 1998, only genotype C2 has been found.
Measles viruses from sporadic cases isolated in France during 2004 were genotyped. Of the three samples collected, two were genotype B3.1. An imported case (a sample from a patient who had recently traveled to Algeria) was isolated in Lyon. The second genotype B3.1 case isolated in Caen was also an imported case, as the patient was in contact with a foreigner. The third sample, isolated from a child 15 days after measles-mumps-rubella vaccination, belonged to genotype A.
Design of the RT-ARMS PCR assay for genotype identification.
The 450 C-terminal nucleotides of the N gene of measles virus contain sufficient nucleotide variation to allow discrimination among genotypes by phylogenetic analysis. This region was selected for the development of an assay that could discriminate between different genotypes by using primers that hybridize with genotype-specific nucleotides, also called SNPs. These SNPs were selected after multiple alignments of different measles virus nucleoprotein C-terminal sequences available from the NCBI database. We designed specific primers that could discriminate between genotypes A, B2, B3.1, B3.2, C2, and D7 (Table 3).
TABLE 3.
Genotype-specific primers for RT-ARMS PCR
| Primer namea | Target genotype | Position, primer sequence (5′-3′)b | Product size (bp) |
|---|---|---|---|
| Warms As | A | nt 1574-1596*, 5′-GAAGGTCAGCTGACGCCCTGgaT-3′ | 104 |
| Warms Aas | nt 1644*-1675, 5′-GATTTCTGTCATTGTACACatT-3′ | ||
| Warms B2s | B2 | nt 1400-1422*, 5′-GATTGGGGGGCAAGGAGGATtcA-3′ | 125 |
| Warms B2as | nt 1500*-1522, 5′-GTGTGCCGGTTGGAAGATGccTG-3′ | ||
| Warms B3.1s | B3.1 | nt 1417-1439*, 5′-GACAGGAGGGTCAAACAGtcC-3′ | 196 |
| Warms B3.1as | nt 1587*-1608, 5′-GGCTTGCAGCCTAAGCAGGcgA-3′ | ||
| Warms B3.2s | B3.2 | nt 1417-1439*, 5′-GAGGACAGGAGGGTCAAACtcG-3′ | 119 |
| Consensus B3.2as | nt 1510-11528, 5′-CTAGGGGTGTGCCGGTTGG-3′ | ||
| Warms C2s | C2 | nt 1292-1314*, 5′-CAGAGATTGCAATGCATACTtgA-3′ | 185 |
| Warms C2as | nt 1450*-1472, 5′-GCCCGGTTTCTCTGTAGCTCagT-3′ | ||
| Consensus D7s | D7 | nt 1442-1464, 5′-GAGAAGCCAGGGAGAGCTACAGAG | 155 |
| Warms D7as | nt 1572*-1594, 5′-GCAGGGCGTCAGCTGACCGTgcG-3′ |
The teminal “a” or “as” in the primer name indicates the plus or the minus sense of the gene transcription, respectively.
The mismaches introduced in the RT-ARMS primers are shown in lowercase letters. * indicates the SNP position according to the assignment described previously by Cattaneo et al. (5). nt, nucleotides.
Genotype-specific reactivity of specific primers in RT-ARMS PCR.
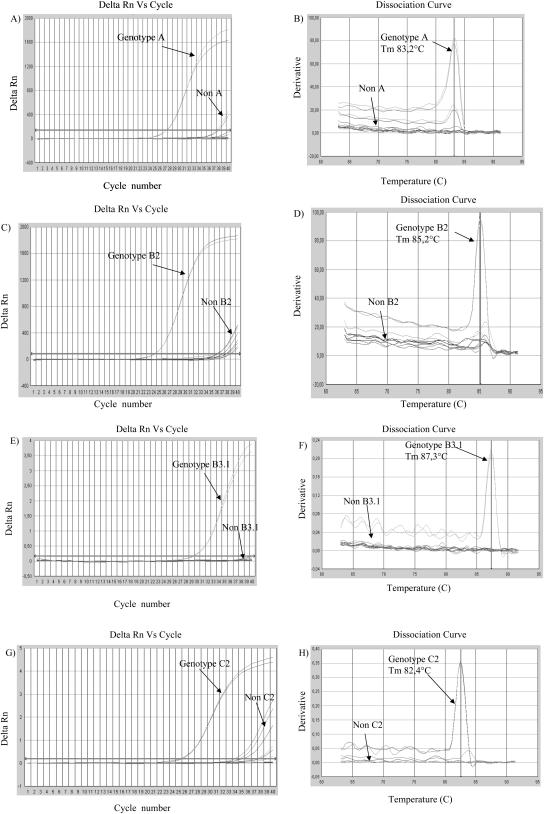
A well-characterized panel of 14 isolates from genotypes A (four isolates), B2 (two isolates), B3.1 (two isolates), B3.2 (two isolates), C2 (two isolates), and D7 (two isolates) was tested with our genotype identification assay. Figures 2 and 3 show the results using the specific genotype primers. Whereas a positive genotype had a cot value of 15 to 30 cycles, the heterotypic genotypes failed to reach a positive value.
FIG. 2.
Genotyping experiment based on RT-ARMS PCR using SYBR green. Different samples of MV genotypes A, B2, B3.1, B3.2, C2, and D7 were tested. The amplification curves of the isolates that reacted with primers specific for genotype A (A), genotype B2 (C), genotype B3.1 (E), and genotype C2 (G) are indicated, including a representative nonreactive isolate from each genotype and a no-template reaction. On the corresponding dissociation curves (B, D, F, and H), the melting temperature (Tm) for each genotype has been indicated with a vertical line. Delta Rn, relative fluorescence.
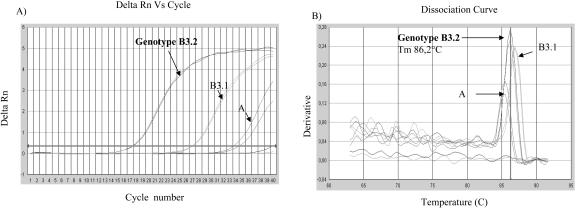
FIG. 3.
Amplification curve (A) of various samples of genotypes A, B2, B3.1, B3.2, C2, and D7 as detected by the genotype B3.2-specific primers. On the dissociation curve (B), the melting-point peaks were clearly separated between genotypes A, B3.1, and B3.2. Delta Rn, relative fluorescence.
In order to analyze the sensitivity of the RT-ARMS PCR, three different concentrations were used per specimen. Good amplification curves were obtained with 0.26 ng of PCR product, whereas higher concentrations led to the inhibition of the RT-ARMS PCR. We therefore used 0.26 ng of target in all assays described in this study. All PCR products amplified after 32 cycles were considered to be false positives.
The primers designed to detect genotypes A, B2, B3.1, and C2 were highly specific (Fig. 2), as only the targeted genotype was amplified by the corresponding primers. The Tm values for genotypes A, B2, B3.1, and C2 were 83.2, 85.2, 87.3, and 82.4°C, respectively.
The primers for genotype B3.2 reacted preferentially with the targeted genotype; however, the B3.1 genotype was also amplified. In this case, the B3.2-specific primers were a hundred times less efficient than in the homologous system. However, the PCR products from different genotypes could be differentiated by their different melting temperatures (Fig. 3B). The genotype B3.2 product had a Tm value of 86.2°C, whereas non-B3.2 genotypes amplified by the B3.2-specific primers had Tm values of 85.4 and 87.1°C for genotypes A and B3.1, respectively. Similar results, i.e., different peaks but distinguished by Tm values, were obtained with the primers for genotype D7 (data not shown).
Measles outbreak investigations by RT-ARMS PCR.
We tested a panel of 30 samples collected during various measles outbreaks: Gabon in 1984 (3 samples), Cameroon in 2001 (17 samples), Morocco in 2003 (7 samples), and sporadic cases in France in 2004 (3 samples). These viruses included the genotypes A (1 sample), B2 (3 samples), B3.1 (19 samples), and C2 (7 samples), as determined by sequencing and phylogenetic analysis of the C-terminal part of the nucleoprotein (Fig. 1 and 4).
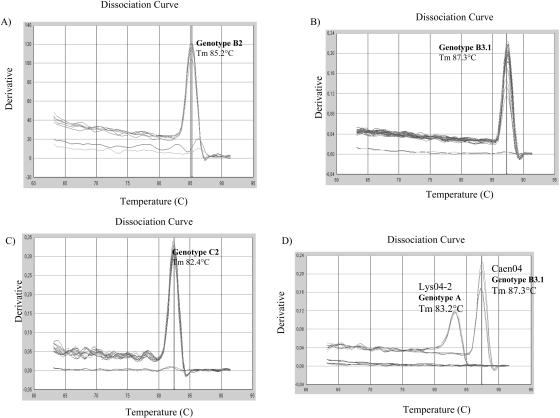
FIG. 4.
Measles outbreak investigation by RT-ARMS PCR. Representative results for genotyping of 30 samples isolated during measles outbreaks in Gabon in 1984 (A), Cameroon in 2001 (B), Morocco in 2003 (C), and France in 2004 (D) are shown. The genotype and the corresponding Tm are indicated. For each outbreak, the melting curve analysis includes a positive control and a no-template sample.
The sample of genotype A was positive with the genotype A-specific primers, with a Tm of 83.2°C corresponding to the specific Tm of genotype A. All three B2 and seven C2 samples were positive with B2- and C2-specific primers with corresponding Tm values of 85.2 and 82.4°C, respectively. Of the 19 samples of genotype B3.1, all but one were positive with genotype B3.1-specific primers, with a Tm of 87.3°C specific for genotype B3.1. The sequence of the target region of the single negative virus isolate was analyzed for mismatches with the primer-hybridizing region. The results show that this sample has one mutation, C1611T, next to the mismatches that we introduced in the reverse primer, increasing the number of mismatches to three consecutives nucleotides, between the target sequence and the specific primer, thus probably prohibiting hybridization of the primer.
To confirm the specificity of the RT-ARMS PCR method, at least three samples from each outbreak were investigated using all sets of primers designed in this study. The results show that only the A specimens were amplified by the genotype A-specific primers. Similar results were observed with B2-, B3.1-, and C2-specific primers, which specially amplified the corresponding specimens. In contrast, primers specific for genotypes B3.2 and D7 amplified at least two other genotypes but with a lower efficacy than their corresponding targets (data not shown).
DISCUSSION
We have developed an RT-ARMS PCR assay to genotype measles virus. The primers used for this assay were based on the C-terminal sequence of the N gene, which is the most variable region of MV genome. The success of the RT-ARMS PCR method relies on the detection of a genotype-specific nucleotide by the designed primers. However, our results show that the specificity of the reaction is enhanced when the two primers (forward and reverse) have an SNP. Nevertheless, even with only one specific primer, different genotypes could be differentiated by analysis of their melting curves. We observed this result when primers specific for genotypes B3.2 and D7 were used. The differentiation between different genotypes using the same pair of primers is explained by the fact that the Tm value is directly related to the nucleotide sequence variation (15, 20). The analysis of the nucleotide composition of the fragment amplified with genotype B3.2-specific primers showed an average percentage of GC content of 63.7 and 63.1% for genotype B3.1 and B3.2, respectively. Despite the fact that the GC contents of these genotypes are relatively close, these two genotypes could be differentiated, indicating the sensitivity of our method.
Different studies have been developed for virus genotyping by real-time PCR. Plantier et al. (16) developed a real-time PCR for human immunodeficiency virus genotyping, and Bruce et al. (3) developed a real-time PCR for specific detection of the RV2 lineage of rhadinoviruses, but all these studies used a fluorescent probe to enhance the specificity of the reaction. In an attempt to increase the specificity of the RT-ARMS primer, we deliberately introduced mismatches near the 3′ ends. These primers allowed specific amplification of thetarget sequence followed by SYBR green binding and monitoring of PCR product accumulation. It therefore eliminated the need to synthesize expensive fluorescent probes. Furthermore, this approach can be used to efficiently enhance specificity in other hybridization protocols.
The feasibility of RT-ARMS genotyping of MV was demonstrated using samples from different epidemics. Rapid investigation of measles outbreaks provides important information about vaccination program impact and ensures the implementation of appropriate outbreak response activities. Data on the MV genotype in Gabon in 1984 confirmed that only the B2 genotype was circulating in that region. Since 1984, genotype B2 has not been found elsewhere. With the implementation of MV surveillance by the WHO in African countries, further molecular epidemiology studies will clarify whether or not the B2 genotype is still circulating in Central Africa.
The analysis of isolates from Cameroon in 2001 show that only genotype B3.1 was found. Genotype B1 was identified in Cameroon in 1983 (19) and has not been found since. It appears to have been replaced by genotype B3.1 (9), or it may be present in areas that have not been investigated.
In Morocco, only genotype C2 was found during the 2003 epi-demic. An imported case of MV of genotype C2 from Morocco was found during the same year in Spain (12). Since the beginning of molecular characterization of MV in Morocco in 1998 (1), only genotype C2 has been identified. It probably represents the endemic strain in that country.
In France during 2004, sporadic measles cases were genotype A (a postvaccination case) and genotype B3.1, genotypes imported from Africa. We previously reported that MV genotype B3.1 was geographically restricted to Central Africa and the Sudan (9). The advent of modern travel has led to a large distribution of these viruses. The importation of MV genotype B3.1 from Africa in Spain during the 2003 epidemic was previously reported (12). The first case of African MV imported to France was isolated in Lyon in 1994 and belonged to genotype B3.2 (7). The number of African measles importations in Europe is increasing, and this raises the question of the coordination of MV molecular surveillance between the two continents.
In this study, we described a real-time ARMS PCR assay for genotyping of six measles genotypes. Using the results obtained by direct sequencing as a reference, the data show that the accuracy of MV genotyping by melting curve analysis was approximately 97%. Despite similar reproducibility, the RT-ARMS PCR offers several advantages over sequencing analysis. First, the RT-ARMS PCR technique is more sensitive, as only 0.26 ng of the PCR product is required instead of the 50 ng required for the sequencing technique. Second, the time saved by this approach is considerable. The throughput in the laboratory has increased from approximately 3 days for 30 genotypes by sequencing and phylogenetic analysis to 2 h by RT-ARMS PCR assay. Finally, the RT-ARMS PCR genotyping assay is more appropriate for high-throughput screening, as it does not required extensive post-PCR manipulation.
In conclusion, the RT-ARMS PCR assay showed good genotyping accuracy and was more sensitive and rapid. The real-time PCR technology is presently available in some laboratories in developing countries (16). Therefore, our method could be a useful and simple tool for molecular epidemiological studies by allowing rapid discrimination between different genotypes in field conditions.
Acknowledgments
The studies were supported by the Direction Générale de la Santé, France, and INSERM.
REFERENCES
- 1.Alla, A., S. L. Liffick, B. R. Newton, R. Elaouad, P. A. Rota, and W. J. Bellini. 2002. Genetic analysis of measles viruses isolated in Morocco. J. Med. Virol. 68:441-444. [DOI] [PubMed] [Google Scholar]
- 2.Bai, R. K., and L. J. Wong. 2004. Detection and quantification of heteroplasmic mutant mitochondrial DNA by real-time amplification refractory mutation system quantitative PCR analysis: a single-step approach. Clin. Chem. 50:996-1001. [DOI] [PubMed] [Google Scholar]
- 3.Bruce, A. G., A. M. Bakke, M. E. Thouless, and T. M. Rose. 2005. Development of a real-time QPCR assay for the detection of RV2 lineage-specific rhadinoviruses in macaques and baboons. Virol. J. 2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckland, R., C. Gerald, D. Barker, and F. Wild. 1988. Cloning and sequencing of the nucleoprotein gene of measles virus (Halle strain). Nucleic Acids Res. 16:11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cattaneo, R., A. Schmid, P. Spielhofer, K. Kaelin, K. Baczko, V. ter Meulen, J. Pardowitz, S. Flanagan, B. K. Rima, S. A. Udem, et al. 1989. Mutated and hypermutated genes of persistent measles viruses which caused lethal human brain diseases. Virology 173:415-425. [DOI] [PubMed] [Google Scholar]
- 6.Combredet, C., V. Labrousse, L. Mollet, C. Lorin, F. Delebecque, B. Hurtrel, H. McClure, M. B. Feinberg, M. Brahic, and F. Tangy. 2003. A molecularly cloned Schwarz strain of measles virus vaccine induces strong immune responses in macaques and transgenic mice. J. Virol. 77:11546-11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fayolle, J., B. Verrier, R. Buckland, and T. F. Wild. 1999. Characterization of a natural mutation in an antigenic site on the fusion protein of measles virus that is involved in neutralization. J. Virol. 73:787-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 9.Kouomou, D. W., E. Nerrienet, J. Mfoupouendoun, G. Tene, H. Whittle, and T. F. Wild. 2002. Measles virus strains circulating in Central and West Africa: geographical distribution of two B3 genotypes. J. Med. Virol. 68:433-440. [DOI] [PubMed] [Google Scholar]
- 10.Kouomou, D. W., and T. F. Wild. 2002. Adaptation of wild-type measles virus to tissue culture. J. Virol. 76:1505-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kremer, J. R., F. Fack, C. M. Olinger, M. N. Mulders, and C. P. Muller. 2004. Measles virus genotyping by nucleotide-specific multiplex PCR. J. Clin. Microbiol. 42:3017-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosquera, M. M., F. Ory, and J. E. Echevarria. 2005. Measles virus genotype circulation in Spain after implementation of the national measles elimination plan 2001-2003. J. Med. Virol. 75:137-146. [DOI] [PubMed] [Google Scholar]
- 13.Newton, C. R., A. Graham, L. E. Heptinstall, S. J. Powell, C. Summers, N. Kalsheker, J. C. Smith, and A. F. Markham. 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17:2503-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parks, C. L., R. A. Lerch, P. Walpita, H. P. Wang, M. S. Sidhu, and S. A. Udem. 2001. Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J. Virol. 75:910-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payungporn, S., P. Tangkijvanich, P. Jantaradsamee, A. Theamboonlers, and Y. Poovorawan. 2004. Simultaneous quantitation and genotyping of hepatitis B virus by real-time PCR and melting curve analysis. J. Virol. Methods 120:131-140. [DOI] [PubMed] [Google Scholar]
- 16.Plantier, J. C., M. Gueudin, F. de Oliveira, F. Damond, V. Lemee, F. Brun-Vezinet, and F. Simon. 2004. Rapid discrimination between human immunodeficiency virus type 2 groups A and B by real-time PCR. J. Clin. Microbiol. 42:5866-5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuel, D., S. Beard, H. Yang, N. Saunders, and L. Jin. 2003. Genotyping of measles and mumps virus strains using amplification refractory mutation system analysis combined with enzyme immunoassay: a simple method for outbreak investigations. J. Med. Virol. 69:279-285. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi, M., T. Nakayama, Y. Kashiwagi, T. Takami, S. Sonoda, T. Yamanaka, H. Ochiai, T. Ihara, and T. Tajima. 2000. Single genotype of measles virus is dominant whereas several genotypes of mumps virus are co-circulating. J. Med. Virol. 62:278-285. [PubMed] [Google Scholar]
- 19.Taylor, M. J., E. Godfrey, K. Baczko, V. ter Meulen, T. F. Wild, and B. K. Rima. 1991. Identification of several different lineages of measles virus. J. Gen. Virol. 72:83-88. [DOI] [PubMed] [Google Scholar]
- 20.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-131, 134-138. [DOI] [PubMed] [Google Scholar]
- 21.World Health Organization. 2005. Measles. Fact sheet no. 286. World Health Organization, Geneva, Switzerland.
- 22.World Health Organization. 2003. Update of the nomenclature for describing the genetic characteristics of wild-type measles viruses: new genotypes and reference strains. Wkly. Epidemiol. Rec. 78:229-232. [PubMed] [Google Scholar]
- 23.Zandotti, C., D. Jeantet, F. Lambert, D. Waku-Kouomou, F. Wild, F. Freymuth, J. R. Harle, X. de Lamballerie, and R. N. Charrel. 2004. Re-emergence of measles among young adults in Marseilles, France. Eur. J. Epidemiol. 19:891-893. [DOI] [PubMed] [Google Scholar]