Abstract
Mycobacterium tuberculosis strains of the Beijing genotype were first identified in China and neighboring countries and have attracted special attention due to their global emergence and association with drug resistance. To further analyze the spread and special characteristics of Beijing genotype strains, accurate, rapid and sensitive methods that overcome the drawbacks of the classical methods such as IS6110 DNA fingerprinting or spoligotyping for the identification of strains of this genotype are needed. Based on the nucleotide sequences of M. tuberculosis SAWC0780 and H37Rv, primers and fluorogenic 5′ nuclease (TaqMan) probes for real-time PCR assays specific for Beijing and non-Beijing strains, respectively, were designed. The detection limits for the real-time PCR assays were about 5 and 10 copies of chromosomal DNA, respectively. In mixtures of Beijing and non-Beijing DNA, a multiplex assay was able to detect (i) one copy of Beijing DNA in approximately 1,000 copies of non-Beijing DNA and (ii) one copy of non-Beijing DNA in approximately 2,000 copies of Beijing DNA. In a blinded analysis of a collection of 103 multidrug-resistant strains isolated in Germany in 2001, all 62 Beijing and all 41 non-Beijing strains were correctly identified. In conclusion, the real-time assay allows for the rapid and specific detection of Beijing and non-Beijing strains. The major advantages of this test in comparison to other methods used for the identification of Beijing strains are its simplicity and sensitivity and the fact that amplification and detection occur within one reaction tube.
In recent years the Beijing genotype of Mycobacterium tuberculosis has attracted special attention because of its global emergence (2). The strains were first described in China and neighboring countries in 1995 (12), and subsequently the occurrence of Beijing genotype strains has been documented in several parts of the world, especially in Asian countries but also in the former Soviet Union, Europe, Africa, and the United States (1, 2, 12). The Beijing genotype has caused outbreaks of multidrug-resistant (MDR) tuberculosis (8, 9), and some investigations have indicated an association with drug resistance (2). Additionally, reported differences in the clinical picture of Beijing genotype infection and the immune response it evoked stimulated a discussion on the special characteristics of Beijing genotype strains (6, 7, 10).
Accurate methods for the identification of Beijing genotype strains are a prerequisite for further studies analyzing the spread and the characteristics of the Beijing genotype family. The main methodologies used so far are IS6110 DNA fingerprinting (13) and spoligotyping (3), and recently a consensus definition based on these methods has been published (4). Both techniques, however, involve different working steps in the laboratory, and especially the IS6110 fingerprinting method requires a high level of standardization and normalization procedures to ensure comparable results. Furthermore, the sensitivity of both methods to identify the presence of subpopulations of Beijing or non-Beijing strains, e.g., in the case of mixed infections, or to differentiate Beijing and non-Beijing strains directly in clinical material is low.
Warren and coworkers (14) recently described a very sensitive PCR-based methodology that is easier to perform. They successfully applied two PCRs targeting a specific chromosomal deletion present in Beijing strains to identify a large number of mixed infections with Beijing and non-Beijing strains in sputum samples, a finding with important implications for the understanding of tuberculosis epidemiology. The combination of a very sensitive PCR technology with subsequent analysis of the PCR products by agarose gel electrophoresis, however, is inherently hampered by the latent problem of contamination that could complicate the interpretation of results.
We therefore developed a multiplex real-time PCR assay which allows the specific identification of Beijing and non-Beijing DNA in a one-tube reaction. The real-time PCR assay was then evaluated by analyzing a well-characterized collection of MDR strains from Germany, for which the classification as Beijing and non-Beijing strains has been performed with the “gold standard” methods IS6110 DNA fingerprinting and spoligotyping (5).
MATERIALS AND METHODS
Strains analyzed.
A set of 103 MDR strains obtained from patients living in Germany in 2001 (5) was used to evaluate the capability of the Beijing real-time PCR assay established in this investigation for discrimination of Beijing and non-Beijing genotype strains.
Genotypic characterization.
To allow a precise classification into the two categories Beijing and non-Beijing, all strains were analyzed by IS6110 DNA fingerprinting and spoligotyping as described previously (3, 11). Chromosomal DNA was isolated using a standard procedure as describe elsewhere (11). The genotyping results have been described in detail in our previous investigation (5).
Design of real-time PCR primer and probe sets.
Primers and probes were synthesized by ABI (Applied Biosystems, Weiterstadt, Germany) and TibMolBiol (Berlin, Germany), respectively.
Real-time PCR assays.
Real-time PCR experiments were run with the Rotor-Gene 2000 (Corbett Research, Mortlake, Australia) and ABI TaqMan Universal PCR Master Mix under reaction conditions of 95° for 10 min and 40 to 50 two-step cycles consisting of 92°C for 15 s and 60°C for 1 min. The primer concentrations in the reaction mixtures of 15 μl were 500 nM for each forward and reverse primer and 200 nM for each probe. A total of 1.5 μl of a 1:100 dilution of chromosomal DNA was used as a PCR template.
RESULTS AND DISCUSSION
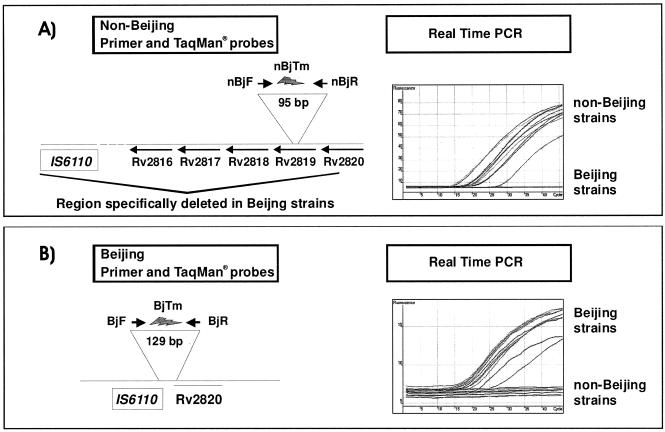
Based on the nucleotide sequences of GenBank accession numbers BX842581 and AF390039, primers and fluorogenic 5′ nuclease (TaqMan) probes for real-time PCR assays specific for Beijing and non-Beijing strains, respectively, were designed (Table 1). (Primers and probes were synthesized by ABI and TibMolBiol, respectively.) Analysis with BLASTN demonstrated that hypothetical DNA fragments generated with primers nBjF/nBjR (non-Beijing) and BjF/BjR (Beijing) are identical with a fragment of M. tuberculosis H37Rv or M. tuberculosis SAWC0780 (Beijing), respectively. The detection of the distinct PCR fragments is based on specific TaqMan probes labeled with different fluorescent dyes. Probe BjTM that binds to the 129-bp fragment generated by the primers nBjF/nBjR (Table 1 and Fig. 1) was labeled with a 5′ FAM (6-carboxyfluorescein) label and a 3′ TAMRA (6-carboxytetramethylrhodamine) quencher. Conversely, nBjTM that binds to the 95-bp fragment generated by primers nBjF/nBjR with a 5′ Yakima Yellow label and a 3′ DABCYL [4-(4′-dimethylaminophenylazo)benzoic acid] quencher (Table 1 and Fig. 1).
TABLE 1.
Sequences of primers and TaqMan probes used for detection of Beijing and non-Beijing type strains
| Primers and probe for target strains | Name | Sequence |
|---|---|---|
| Non-Beijing strainsa | ||
| Forward primer | nBjF | 5′-AAGCATTCCCTTGACAGTCGAA-3′ |
| Fluorogenic probe | nBjTM | 5′-6FAM-TCCAAGGTCTTTG-MGB-NFQ-3′c |
| Reverse primer | nBjR | 5′-GGCGCATGACTCGAAAGAAG-3′ |
| Beijing strainsb | ||
| Forward primer | BjF | 5′-CTCGGCAGCTTCCTCGAT-3′ |
| Fluorogenic probe | BjTM | 5′-YAK-AACGCCAGAGACCAGCCGCCGGCT-DB-3′d |
| Reverse primer | BjR | 5′-CGAACTCGAGGCTGCCTACTAC-3′ |
The primers and probe for non-Beijing strains were based on GenBank accession no. BX842581, nucleotides 7896 to 7990. The fragment generated by the primers was 95 bp in length.
The primers and probe for Beijing strains were based on GenBank accession no. AF390039, nucleotides 820 to 948. The fragment generated by the primers was 129 bp in length.
MGB-NFQ, minor groove binding, nonfluorescent quencher.
YAK-DB, Yakima yellow-DABCYL (nonfluorescent dark quencher).
FIG. 1.
Schematic localization of primers and probes and PCR results of 10 randomly chosen Beijing and non-Beijing strains of the established Beijing and non-Beijing real-time PCR assays.
The performance of the real-time PCRs and the specificity of the primer-probe sets were initially tested by analysis of 10 randomly chosen Beijing and 10 non-Beijing strains, classified by IS6110 DNA fingerprinting and spoligotyping. With an annealing temperature of 60°C and a maximum number of 50 PCR cycles, both real-time assays yielded only fluorescence when DNA from Beijing genotype strains or non-Beijing strains was analyzed, respectively (Fig. 1). No cross-reactivity of the Beijing real-time PCR assay for non-Beijing DNA or, vice versa, of the non-Beijing real-time PCR assay for Beijing DNA was observed. The identity of the PCR products with the respective genomic fragments was confirmed by DNA sequencing also (data not shown).
To estimate the detection limit of both PCR assays, dilution series of chromosomal DNA (over seven orders of magnitude) of the Beijing and non-Beijing strains were PCR amplified. Absolute gene copy numbers were estimated based on spectrophotometric measurement of DNA concentration (UV absorption at 260 nm) and the assumption that one genome of M. tuberculosis has a molecular weight of 4 fg. Standard curves for each target sequence with defined quantities of DNA were obtained for each experiment (using a 1:10 dilution series, beginning with 107 copies), consistently providing significant correlations (r2 values always of >0.99; data not shown). The sensitivity of both PCRs was very high, since the detection limits of the PCRs could be calculated to 5 and 10 copies of chromosomal DNA for the non-Beijing PCR and the Beijing PCR, respectively.
Since the real-time PCR machine is able simultaneously to measure multiple dyes with distinct emission wavelengths, a multiplex PCR assay applying both PCRs in one reaction was established. To test whether the multiplex PCR worked properly, primers and probes were tested by analyzing 10 DNA samples containing either Beijing or non-Beijing DNA and 10 samples with mixtures of non-Beijing and Beijing DNA (ratio of 1:1 to 1:100). In all cases, clear signals in the respective fluorescence detecting channels were obtained when Beijing or non-Beijing DNA was present only, while when mixtures of both DNA types were analyzed, signals in both channels were obtained (data not shown). Thus, no cross-reaction of the Beijing PCR probe system for non-Beijing DNA or of the non-Beijing PCR probe system for Beijing DNA was observed.
The detection limit of the multiplex PCR for Beijing and non-Beijing DNA was then determined by analyzing DNA samples containing a fixed amount of Beijing DNA (106 copies) mixed with DNA from a 1:10 dilution series of non-Beijing DNA (beginning with 106 copies) and vice versa. Accordingly, the detection limits of the multiplex real-time PCR assay were calculated to (i) one copy of Beijing DNA in approximately 1,000 copies of non-Beijing DNA and (ii) one copy of non-Beijing DNA detected in approximately 2,000 copies of Beijing DNA.
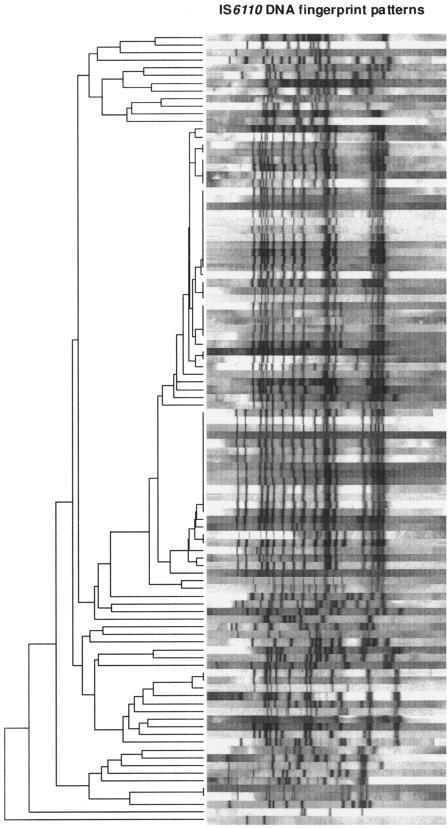
The capability of the multiplex PCR assay for the rapid identification of clinical Beijing and non-Beijing strains was tested by blinded analyses of a collection of 103 MDR strains isolated in Germany in 2001 for which stratification into Beijing and non-Beijing strains had already performed based on spoligotyping and IS6110 DNA fingerprinting data (5). The population structure based on IS6110 DNA fingerprinting is shown in Fig. 2. Based on identical IS6110 DNA fingerprint patterns, 47 of the 103 isolates were grouped in 10 different clusters comprised of from 2 to 14 isolates. The multiplex real-time assay correctly identified 62 isolates as Beijing genotype and 41 isolates as non-Beijing genotype. No false-positive signals were observed for Beijing and non-Beijing strains, although the PCR was proven to be highly sensitive. Overall, a 100% concordance between the new real-time PCR and genotyping data based on spoligotyping and IS6110 DNA fingerprinting was obtained. Thus, the specificity and sensitivity of the multiplex PCR assay for the detection of Beijing and non-Beijing strains were calculated to be 100% in this investigation.
FIG. 2.
IS6110 DNA fingerprint patterns of the 103 MDR strains analyzed. The IS6110 banding patterns are ordered by similarity in a dendrogram. The position of each IS6110 band is normalized so that banding patterns of all strains are mutually comparable. The scale depicts similarity of IS6110 patterns calculated with the Dice coefficient and the unweighted-pair group method using average linkages method.
The established real-time PCR assay allows for the rapid and sensitive detection of Beijing and non-Beijing strains in a one-tube reaction without any further working steps. The simplicity of this test allows for analysis of samples on various real-time PCR machines. The major advantages of this test in comparison to other methods used for identification of Beijing strains are its simplicity and sensitivity and the fact that amplification and detection occur within one reaction tube.
In contrast, the “gold standard” IS6110 DNA fingerprint method requires several working steps, a well-grown bacterial culture, and analysis with 19 M. tuberculosis Beijing reference strains when optimal classification is required (4). In addition, IS6110 DNA fingerprinting must be performed by applying a highly standardized methodology and also involves analysis with a sophisticated software program allowing normalization of restriction fragment length polymorphism patterns (11). The PCR-based techniques described so far, spoligotyping (3) and the method described by Warren et al. (14), allow much easier detection of Beijing strains; however, in both cases further analysis of PCR products is necessary. Using the latter method, these additional working steps may pose a high risk of contamination, especially when the detection of small amounts of DNA in clinical samples is desired. This potential risk can be completely avoided by the established real-time assay, which has a further advantage that it allows quantification of the relative amounts of Beijing and non-Beijing DNA in a given sample. The high sensitivity and specificity in combination with the simplicity of the method make the Beijing and non-Beijing real-time PCR assay a very useful tool that will facilitate improved studies of different aspects of the M. tuberculosis Beijing genotype in the future.
Acknowledgments
The authors thank I. Radzio, B. Schlüter, T. Ubben, and P. Vock for excellent technical assistance.
Parts of this work were supported by the Robert-Koch Institute, Berlin, Germany.
REFERENCES
- 1.Bifani, P. J., B. Mathema, N. E. Kurepina, B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 2.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kremer, K., J. R. Glynn, T. Lillebaek, S. Niemann, N. E. Kurepina, B. N. Kreiswirth, P. J. Bifani, D. van Soolingen. 2004. Definition of the Beijing/W lineage of Mycobacterium tuberculosis on the basis of genetic markers. J. Clin. Microbiol. 42:4040-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubica, T., S. Rüsch-Gerdes, S. Niemann. 2004. The Beijing genotype is emerging among multidrug-resistant Mycobacterium tuberculosis strains from Germany. Int. J. Tuberc. Lung Dis. 8:1107-1113. [PubMed] [Google Scholar]
- 6.Lopez, B., D. Aguilar, H. Orozco, M. Burger, C. Espitia, V. Ritacco, L. Barrera, K. Kremer, R. Hernandez-Pando, K. Huygen, D. van Soolingen. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manabe, Y. C., A. M. Dannenberg Jr., S. K. Tyagi, C. L. Hatem, M. Yoder, S. C. Woolwine, B. C. Zook, M. L. Pitt, W. R. Bishai. 2003. Different strains of Mycobacterium tuberculosis cause various spectrums of disease in the rabbit model of tuberculosis. Infect. Immun. 71:6004-6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moss, A. R., D. Alland, E. Telzak, D. Hewlett Jr., V. Sharp, P. Chiliade, V. LaBombardi, D. Kabus, B. Hanna, L. Palumbo, K. Brudney, A. Weltman, K. Stoeckle, K. Chirgwin, M. Simberkoff, S. Moghazeh, W. Eisner, M. Lutfey, B. Kreiswirth. 1997. A city-wide outbreak of a multiple-drug-resistant strain of Mycobacterium tuberculosis in New York. Int. J. Tuberc. Lung Dis. 1:115-121. [PubMed] [Google Scholar]
- 9.Narvskaya, O., T. Otten, E. Limeschenko, N. Sapozhnikova, O. Graschenkova, L. Steklova, A. Nikonova, M. L. Filipenko, I. Mokrousov, B. Vyshnevskiy. 2002. Nosocomial outbreak of multidrug-resistant tuberculosis caused by a strain of Mycobacterium tuberculosis W-Beijing family in St. Petersburg, Russia. Eur. J. Clin. Microbiol. Infect. Dis. 21:596-602. [DOI] [PubMed] [Google Scholar]
- 10.van Crevel, R., R. H. Nelwan, W. de Lenne, Y. Veeraragu, A. G. van der Zanden, Z. Amin, J. W. van der Meer, D. van Soolingen. 2001. Mycobacterium tuberculosis Beijing genotype strains associated with febrile response to treatment. Emerg. Infect. Dis. 7:880-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Soolingen, D., K. Kremer, E. Vynycky. 2003. New perspectives in the molecular epidemiology of tuberculosis, p. 17-45. In S. H. E. Kaufmann and H. Hahn (ed.), Mycobacteria and TB, vol. 2. Karger, Basel, Switzerland. [Google Scholar]
- 14.Warren, R. M., T. C. Victor, E. M. Streicher, M. Richardson, N. Beyers, N. C. van Pittius, P. D. van Helden. 2004. Patients with active tuberculosis often have different strains in the same sputum specimen. Am. J. Respir. Crit. Care Med. 169:610-614. [DOI] [PubMed] [Google Scholar]