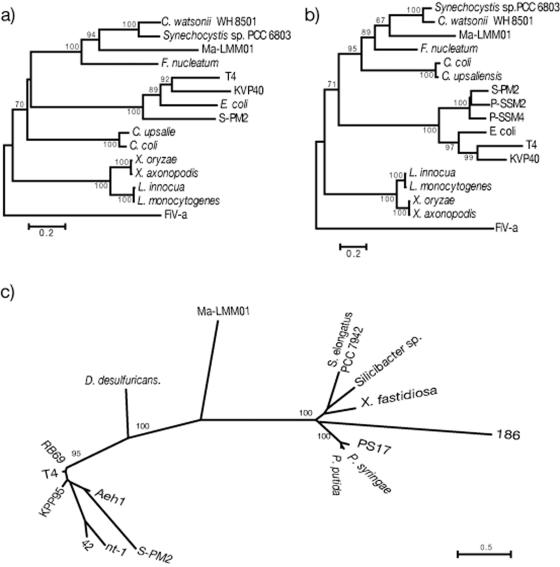
FIG. 6.
Phylogenetic neighbor-joining trees calculated from confidently aligned regions of the amino acid sequences of ribonucleotide reductase alpha-subunit (a) and beta-subunit (b) and putative sheath protein (c) of Ma-LMM01. In panels a and b, the corresponding amino acid sequences of Feldmannia irregularis virus a (FiV-a) were used as outgroups. The phylogenetic tree for the putative sheath protein (c) is unrooted due to the lack of suitable outgroup sequences. Nodes with bootstrap values less than 70% were collapsed. Amino acid sequences of the following organisms were used in the phylogenetic analyses (accession numbers are NCBI accession numbers unless indicated otherwise): for the ribonucleotide reductase alpha-subunit, Synechocystis sp. strain PCC6803 (accession number NP_441654), Crocosphaera watsonii WH8501 (ZP_00179687), Fusobacterium nucleatum subsp. nucleatum ATCC 25586 (ZP_00144798), Listeria monocytogenes EGD-e (NP_465679), Listeria innocua Clip11262 (NP_471591), Xanthomonas oryzae pv. oryzae KACC10331 (YP_199113), Xanthomonas axonopodis pv. citri strain 306 (AAM38910), Campylobacter upsaliensis RM3195 (ZP_00371289), Campylobacter coli RM2228 (ZP_00368144), Escherichia coli (5R1R_C), enterobacterial phage T4 (AAD42621), bacteriophage S-PM2 (CAF34215), bacteriophage KVP40 (AAQ64346), and Feldmannia irregularis virus a (AAR26844); for the ribonucleotide reductase beta-subunit, Synechocystis sp. strain PCC 6803 (NP_443040), Crocosphaera watsonii WH8501 (ZP_00178432), Fusobacterium nucleatum subsp. nucleatum ATCC 25586 (NP_603013), Listeria monocytogenes EGD-e (NP_465678), Listeria innocua Clip11262 (NP_471590), Xanthomonas oryzae pv. oryzae KACC10331(YP_199114), Xanthomonas axonopodis pv. citri strain 306 (AAM38909), Campylobacter upsaliensis RM3195 (ZP_00371674), Campylobacter coli RM2228 (ZP_00367491), Escherichia coli (1MXR_B), enterobacterial phage T4 (AAD42624), bacteriophage S-PM2 (CAF34216), cyanophage P-SSM2 (AAW48101), cyanophage P-SSM4 (AAW50173), bacteriophage KVP40 (AAQ64347), and Feldmannia irregularis virus a (AAR2684); and for sheath protein and sheath-like protein, Xylella fastidiosa Dixon (ZP_00039823), Pseudomonas syringae pv. tomato strain DC3000 (NP_793179), Pseudomonas putida KT2440 (NP_745203), Desulfovibrio desulfuricans G20 (ZP_00130650), Synechococcus elongatus PCC 7942 (ZP_00163909), Silicibacter sp. strain TM1040 (ZP_00339531), bacteriophage PS17 (BAA05467), enterobacterial phage RB69 (AAP76079), bacteriophage S-PM2 (CAF34166), bacteriophage Aeh1 (AAQ17879), enterobacterial phage T4 (GKBPT4), Vibrio natriegens bacteriophage nt-1 (AAG02027), Burkholderia cepacia bacteriophage 42 (42) (AAG02028), enterobacterial phage 186 (P13332), and bacteriophage KPP95 (AAS46616). The amino acid sequences of Ma-LMM01 ORFs encoding the ribonucleotide reductase alpha- and beta-subunits and putative sheath protein are available from the DDBJ (accession numbers AB242259, AB242260, and AB242261, respectively).