Abstract
E-test, Vitek 2, MicroScan, agar dilution, and disk diffusion were compared for detection of decreased linezolid susceptibility due to 23S rRNA gene G2576T mutation among 32 clinical Enterococcus strains initially reported as intermediate or resistant by E-test alone or Vitek 2 confirmed by E-test. Agar and broth dilution methods were in concordance with PCR detection of the mutation, and disk diffusion was somewhat less sensitive but equally specific.
Linezolid provides high rates of clinical cure and microbiological success in complicated infections due to Enterococcus spp., including vancomycin-resistant Enterococcus faecium (3). However, the emergence of resistance during linezolid treatment has been reported for clinical strains of Enterococcus (1, 2, 7, 9, 12, 13). Clinical resistance to linezolid is associated with a G2576T mutation in domain V of 23S rRNA genes of Enterococcus, and the level of linezolid resistance is directly related to the number of 23S rRNA genes containing this mutation (11, 15). Both laboratory and clinical strains of E. faecium with linezolid MICs of 4 μg/ml have been shown to carry the G2576T mutation (10, 14). Accurate detection by susceptibility testing methods of decreased susceptibility due to G2576T mutation in one or two genes is necessary since this can be a prelude to higher levels of linezolid resistance associated with extensive use of the antibiotic (14).
In this study we compared the performance of five different susceptibility testing methods, E-test, disk diffusion, Vitek 2 system, MicroScan WalkAway broth microdilution, and agar dilution, for the detection of decreased linezolid susceptibility of Enterococcus faecalis and E. faecium due to presence of the G2576T mutation.
Strain selection.
Fourteen clinical strains of E. faecium and five strains of E. faecalis reported by the clinical laboratory as linezolid intermediate or resistant based on MICs determined by the E-test (AB Biodisk, Solna, Sweden) were collected during the period January to August 2004. From August 2004 to April 2005, linezolid susceptibility testing was performed by the clinical laboratory utilizing the automated Vitek 2 system (Vitek Systems; bioMérieux, St. Louis, Mo.). Due to the limitation of Vitek 2 in detection of linezolid intermediate or resistant strains, linezolid MICs of ≥4 μg/ml as determined by the Vitek 2 system obtained with clinical isolates of Enterococcus were confirmed by using the E-test. Two strains of E. faecium and eleven of E. faecalis strains reported as linezolid intermediate or resistant were collected during this period. All strains were identified to the species level by using the Vitek 2 system. When the Vitek 2 system failed to identify the strains, identification was obtained by manual biochemical reactions (8). The strains were recovered from blood, urine, respiratory specimens, and various body fluids and tissues. Three well-characterized strains of linezolid-resistant E. faecium (strains 38-13, 45-24, and 38-42) and one strain of lineozlid-resistant E. faecalis (strain 41-31) were kindly provided by Paul Schreckenberger of the Loyola University Medical Center, Maywood, IL.
Susceptibility testing.
E-test linezolid strips with a concentration gradient corresponding to 0.016 to 256 μg/ml were utilized with Mueller-Hinton agar as described by the manufacturer (AB Biodisk, Piscataway, N.J.). E-test MICs were determined as 80% growth inhibition, and measured E-test MICs were rounded up to the next twofold dilution for the categorical interpretation. Disk diffusion testing was performed with 30-μg linezolid disks (BBL, Becton Dickinson) using Clinical and Laboratory Standards Institute (CLSI; formerly National Committee for Clinical Laboratory Standards) standards (4), and 80% growth inhibition was utilized to measure diameters of growth inhibition zones. Agar dilution testing (0.5, 1, 2, 4, and 8 μg/ml) was conducted in accordance with CLSI standards (5) using a linezolid preparation obtained from the manufacturer (lot LZD05003; Pfizer, Inc., Groton, Conn.). Automated susceptibility testing by the Vitek 2 system using the AST GP-61 card (bioMérieux) and the MicroScan WalkAway utilizing the PC 21 plate (Dade Behring, Inc., West Sacramento, Calif.) was performed according to the manufacturers' instructions. The categorical interpretation of results was based on CLSI guidelines (6).
Detection of the G2576T mutation.
To amplify the region of the 23S rRNA gene containing the G2576T mutation, PCR was performed using the following two primers: 5′-GCAGAAGGGAGCTTGACTGCGAG-3′ and 5′-ACCCAGCAATGCCCTTGGCAG-3′ (11). Amplification products were digested with MaeI at 45°C for 4 h and separated by 10% polyacrylamide gel electrophoresis. The G2576T mutation results in addition of a MaeI restriction site (11) and the presence of two DNA bands in polyacrylamide gel electrophoresis.
Findings.
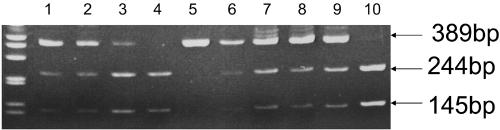
Four of the eight strains of Enterococcus reported by the clinical laboratory as linezolid resistant demonstrated the presence of the G2576T mutation, as well as 1 of the 24 strains clinically reported as linezolid intermediate (Fig. 1).
FIG. 1.
MaeI digestion of PCR product amplified from domain V of the 23S rRNA gene of E. faecium and E. faecalis for presence of the G2576T mutation. Positive control strains are shown in lanes 1 to 4. Lanes 5 to10 show linezolid susceptible (lane 5), intermediate (lanes 6, 8, and 9), and resistant (lanes 7 and 10) clinical strains determined by manual agar dilution and MicroScan broth microdilution.
Because of discrepant results obtained by PCR for G2576T mutation and decreased linezolid susceptibility reported by E-test or Vitek 2 combined with E-test, five different susceptibility testing methods (E-test, Vitek 2, MicroScan broth microdilution, agar dilution, and disk diffusion) were compared to PCR results for G2576T mutation. Of the 18 strains determined to be intermediate or resistant by E-test, only 5 were determined to be PCR positive for the G2576T mutation (Table 1). With the Vitek 2 system, 1 E. faecium strain that was PCR positive for G2576T mutation tested as linezolid susceptible, and only 4 of 13 strains that tested as intermediate or resistant were PCR positive for G2576T mutation (Table 1). In contrast to the E-test and Vitek 2 system, linezolid susceptibility results obtained by the automated MicroScan broth microdilution system and manual agar dilution correlated completely with PCR results (Table 1). All 5 Enterococcus strains demonstrating the 23S rRNA gene G2576T mutation by PCR tested as linezolid intermediate or resistant by MicroScan and agar dilution, and all 27 strains without the G2576T mutation by PCR tested as linezolid susceptible. With the disk diffusion method, four of five strains PCR positive for G2576T mutation tested as linezolid resistant, and one strain that tested as susceptible was positive for the mutation (Table 1). All 27 strains without the G2576T mutation tested as linezolid susceptible by disk diffusion.
TABLE 1.
Correlation of linezolid susceptibility results for E. faecium and E. faecalis with the presence or absence of the G2576T 23S rRNA gene mutation
| Test | MIC (μg/ml) or inhibition zone (mm)a | Total no. of strains | No. of strains with G2576T mutation |
|---|---|---|---|
| E-test | 2 (S) | 14 | 0 |
| 4 (I) | 14 | 1 | |
| ≥8 (R) | 4 | 4 | |
| Vitek 2 | 2 (S) | 19 | 1 |
| 4 (I) | 8 | 1 | |
| ≥8 (R) | 5 | 3 | |
| Microscan | ≤2 (S) | 27 | 0 |
| 4 (I) | 3 | 3 | |
| ≥8 (R) | 2 | 2 | |
| Agar dilution | ≤2 (S) | 27 | 0 |
| 4 (I) | 3 | 3 | |
| ≥8 (R) | 2 | 2 | |
| Disk diffusion | >23 (S) | 28 | 1 |
| ≤20 (R) | 4 | 4 |
S, susceptible; I, intermediate; R, resistant.
Pulsed-field gel electrophoresis revealed a unique band pattern (>6-band difference) for each of the five clinical strains of E. faecium or E. faecalis with the G2576T mutation (16).
Conclusions.
Our results confirm that the presence of the G2576T mutation in clinical strains of E. faecium and E. faecalis results in an increase of the MIC for linezolid to ≥4 μg/ml. Only two methods, automated broth microdilution (MicroScan WalkAway) and manual agar dilution, yielded MICs of ≥4 μg/ml for the 5 clinical strains of E. faecium and E. faecalis demonstrating G2576T mutation by PCR and MICs of ≤2 μg/ml for all 27 clinical strains without the G2576T mutation.
Variability in E-test results likely reflects the inherent difficulty in interpretation by visual examination of 80% growth inhibition end points with the E-test method rounding up MICs. Only 1 of 14 strains with an MIC of 4 μg/ml by E-test was PCR positive for the G2576T mutation (Table 1), indicating MICs in the intermediate range were poorly predictive for presence of the G2576T mutation. The four strains with measured E-test MICs of ≥8 μg/ml were positive for G2576T mutation, indicating that E-test MICs in the resistance range reliably indicate decreased susceptibility of Enterococcus toward linezolid. The Vitek 2 system also demonstrated poor correlation of MICs in the susceptible and intermediate range with the presence or absence of the G2576T mutation (Table 1), likely reflecting a lack of validation of the Vitek AST GP-61 card with linezolid-resistant strains of Enterococcus. Disk diffusion testing appears to be somewhat less sensitive than dilution methods for detection of decreased linezolid susceptibility due to G2576T mutation but specific for detection of fully susceptible strains without the G2576T mutation.
Only the G2576T mutation has been detected to date in clinical isolates of Enterococcus with decreased linezolid susceptibility (13). Until more performance information is available with different methods of susceptibility testing, PCR testing for G2576T mutation should be considered for adjunctive use, especially with vancomycin-resistant strains of Enterococcus isolated from normally sterile body fluids and tissue.
REFERENCES
- 1.Auckland, C., L. Teare, F. Cooke, M. E. Kaufman, M. Warner, G. Jones, K. Bamford, H. Ayles, and A. P. Johnson. 2002. Linezolid-resistant enterococci: report of the first isolates in the United Kingdom. J. Antimicrob. Chem. 50:743-746. [DOI] [PubMed] [Google Scholar]
- 2.Bersos, Z., M. Maniati, F. Kontos, E. Petinaki, and A. N. Maniatis. 2004. First report of a linezolid-resistant vancomycin-resistant Enterococcus faecium strain in Greece. J. Antimicrob. Chem. 53:685-686. [DOI] [PubMed] [Google Scholar]
- 3.Birmingham, M. C., C. R. Rayner, A. K. Meagher, S. M. Flavin, D. H. Batts, and J. J. Schentag. 2003. Linezolid for the treatment of multidrug-resistant, gram-positive infections: experience from a compassionate-use program. Clin. Infect. Dis. 36:159-168. [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2003. Performance standards for antimicrobial disk susceptibility tests, 8th ed. Approved standard M2-A8. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 5.Clinical and Laboratory Standards Institute. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 6.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; fifteenth informational supplement. Approved standard M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 7.Dibo, I., S. K. Pillai, H. S. Gold, M. R. Baer, M. Wetzler, J. L. Slack, P. A. Hazamy, D. Ball, C. B. Hsiao, P. L. McCarthy, Jr., and B. H. Segal. 2004. Linezolid-resistant Enterococcus faecalis isolated from a cord blood transplant recipient. J. Clin. Microbiol. 42:1843-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Facklam, R. R., and M. D. Collins. 1989. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 27:731-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzales, R. D., P. C. Schreckenberger, M. B. Graham, S. Kelkar, K. DenBesten, and J. P. Quinn. 2001. Infections due to vancomycin-resistant Enterococcus faecium resistant to linezolid. Lancet 357:1179. [DOI] [PubMed] [Google Scholar]
- 10.Lobritz, M., R. Hutton-Thomas, S. Marshall, and L. B. Rice. 2003. Recombination proficiency influences frequency and locus of mutational resistance to linezolid in Enterococcus faecalis. Antimicrob. Agents Chemother. 47:3318-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall, S. H., C. J. Donskey, R. Hutton-Thomas, R. A. Salata, and L. B. Rice. 2002. Gene dosage and linezolid resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 46:3334-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pai, M. P., K. A. Rodvold, P. C. Schreckenberger, R. D. Gonzales, J. M. Petrolatti, and J. P. Quinn. 2002. Risk factors associated with the development of infection with linezolid- and vancomycin-resistant Enterococcus faecium. Clin. Infect. Dis. 35:1269-1272. [DOI] [PubMed] [Google Scholar]
- 13.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raad, I. I., H. A. Hanna, R. Y. Hachem, T. Dvorak, R. B. Arbuckle, G. Chaiban, and L. B. Rice. 2004. Clinical-use-associated decrease in susceptibility of vancomycin-resistant Enterococcus faecium to linezolid: a comparison with quinupristin-dalfopristin. Antimicrob. Agents Chemother. 48:3583-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruggero, K. A., L. K. Schroeder, P. C. Schreckenberger, A. S. Mankin, and J. P. Quinn. 2003. Nosocomial superinfections due to linezolid-resistant Enterococcus faecalis: evidence for a gene dosage effect on linezolid MICs. Diagn. Microbiol. Infect. Dis. 47:511-513. [DOI] [PubMed] [Google Scholar]
- 16.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]