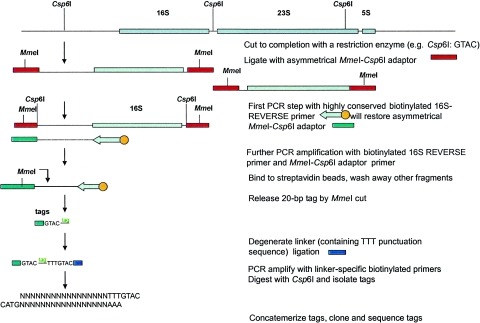
FIG. 1.
Schematic representation of the SP-GST approach on the 16S rRNA gene. Tags are generated upstream of a conserved domain (e.g., position 8 to 27 in the 16S rRNA gene). DNA is first cleaved to completion with Csp6I, the anchoring enzyme. The free cohesive ends are ligated with an asymmetrical oligonucleotide cassette that restores the recognition sequence for the anchoring enzyme and places an MmeI recognition sequence immediately adjacent to the restored sequence. A biotinylated primer specific for the region of position 8 to 27 in the 16S rRNA gene and pointing outward of this gene is used in a first PCR cycle to linearly amplify the region between this specific domain and the most proximal site for the anchoring enzyme. This will result in the synthesis of the complementary strand of the linker fragment. The resulting single-stranded fragment is then exponentially amplified using a primer unique to the restored sequence of the MmeI cassette and the domain-specific primer. The biotinylated products are bound to streptavidin-coated magnetic beads and then digested with MmeI to release the tags, which are further treated as described in our original GST protocol (10).