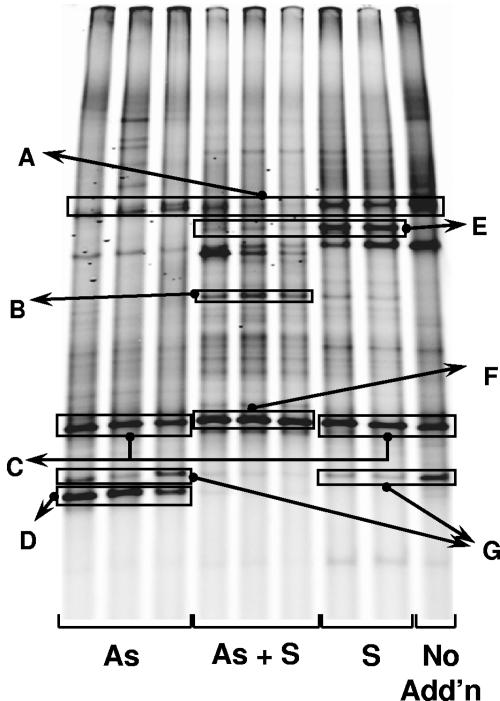
FIG. 6.
DGGE analysis of rrnA amplicons retrieved from various treatments in experiment 1, sampled after 12 days. No Add'n, no additions. DNAs in bands enclosed in boxes were identified either by sequencing or by comparison with the mobilities of cloned fragments. The most similar sequences and similarities were as follows: (A) Picocystis salinarum chloroplast AF454327, >99%; (B) Fusobacteria clone GalB35 AY193168, 94%; (C) mixture of Marinospirillum minutulum AF275713, 95%, and clone ML623J-57, AF507840, >99%; (D) Firmicutes clone un-c23, AJ431345, 90%; (E) Paenibacillus lentimorbus AB110988, 87%; (F) Deltaproteobacteria clone ML623J-57, AF507840, >99%; (G) Desulfuromusa succinoxidans X79415, 95%.