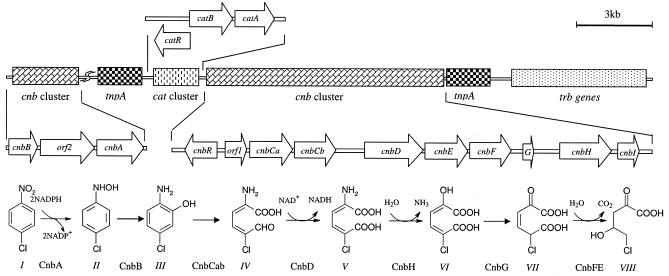
FIG. 1.
Genetic organization of cnb genes involved in 4-chloronitrobenzene (4-CNB) and nitrobenzene degradation and the modified partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1. The genes involved in 4-chloronitrobenzene and nitrobenzene degradation in Comamonas sp. strain CNB-1 are organized in two clusters: a larger cluster (cnbR-orf1-cnbCaCbDEFGHI) includes the putative regulator gene (cnbR), the ring cleavage gene (cnbCab), and several genes of the lower pathway; and a smaller cluster (cnbB-orf2-cnbA) includes genes for the upper pathway that converts 4-chloronitrobenzene to 2-amino-5-chlorophenol and nitrobenzene into 2-aminophenol. The decarboxylation and hydration (from compound VII to compound VIII) was catalyzed by CnbE and CnbF, which was confirmed in recombinant E. coli by simultaneous expression of the two genes (data not shown). Upstream of the genetic cluster cnbR-orf1-cnbCaCbDEFGHI is a genetic cluster, catRBA, that putatively encodes the catechol pathway, and the function of catA, encoding catechol 1,2-dioxygenase, was confirmed by expression in E. coli (data not shown). Between the two cnb genetic clusters, there are some putative genes relating to gene transposition and plasmid conjugation. Arrows indicate the direction of transcription. Symbols: I, 4-chloronitrobenzene; II, 1-hydroxylamino-4-chlorobenzene; III, 2-amino-5-chlorophenol; IV, 2-amino-5-chloromuconic semialdehyde; V, 2-amino-5-chloromuconic acid; VI, 2-oxohex-4-ene-5-chloro-1,6-dioate; VII, 2-oxopent-5-chloro-3-enoate; VIII, 5-chloro-4-hydroxy-2-oxovaletate.