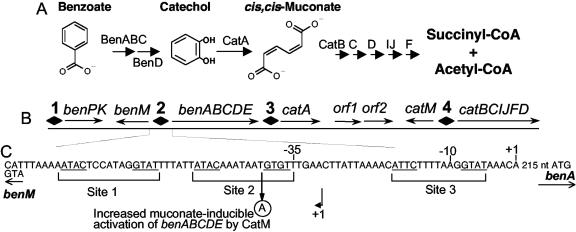
FIG. 1.
BenM and CatM regulate benzoate degradation in ADP1. Catabolism depends on enzymes encoded by the ben and cat genes (A). These chromosomal genes are in an approximately 20 kbp cluster (B) (not drawn to scale). BenM and CatM regulate transcription initiation in four regions (diamonds 1 to 4). The functions of two open reading frames downstream of catA (orf1 and orf2) are unknown, but they are not expressed during growth on benzoate. In the intergenic benMA region (C), there are three potential binding sites for BenM and CatM, as described in the text. Site 1 exactly matches the consensus sequence (underlined) of LysR-type regulators within a subclass to which BenM and CatM belong (10, 31). The sequences in sites 2 and 3 differ from the consensus by a single nucleotide that reduces the extent of dyad symmetry. Above the DNA sequence, the transcription initiation site (+1) and promoter regions (−10 and −35) are shown for benA. The initiation site for the divergently transcribed benM is also indicated (+1 below an arrow). A point mutation (circled A) increases the ability of CatM to activate transcription of the benABCDE operon (12).