Probiotics are defined as “living microorganisms, which upon ingestion in certain numbers exert health benefits on the host beyond inherent basic nutrition” (43). Various studies have indicated that probiotics may alleviate lactose intolerance; have a positive influence on the intestinal flora of the host; stimulate/modulate mucosal immunity; reduce inflammatory or allergic reactions; reduce blood cholesterol; possess anti-colon cancer effects; reduce the clinical manifestations of atopic dermatitis, Crohn's disease, diarrhea, constipation, candidiasis, and urinary tract infections; and competitively exclude pathogens (35, 67, 75, 80, 99). Considering this impressive list of potential health-promoting benefits, it is not surprising that there continues to be considerable interest in the use of probiotics as biotherapeutic agents (67, 75, 82). Furthermore, given a heightened awareness among consumers of the link between diet and health and the fact that probiotic-containing foods are generally perceived as “safe” and “natural,” the global market for such foods is on the increase, particularly dairy-based products marketed for the prophylaxis or alleviation of gastrointestinal disorders (84).
The selection of potential probiotic strains that would be capable of performing effectively in the gastrointestinal tract is a significant challenge. Strain selection has generally been based on in vitro tolerance of physiologically relevant stresses: e.g., low pH, elevated osmolarity, and bile (26, 53, 77, 100). In addition to these physiological assays, molecular investigations are now under way to determine the genetic basis of gastric survival and functionality (11, 78, 79) and inclusion of molecular markers identified by this approach into screening programs may lead to more well-defined and reliable results.
The ability of probiotic strains to hydrolyze bile salts has often been included among the criteria for probiotic strain selection, and a number of bile salt hydrolases (BSHs) have been identified and characterized. However, microbial BSH activity has also been mooted to be potentially detrimental to the human host, and thus it is as yet not completely clear whether BSH activity is in fact a desirable trait in a probiotic bacterium. We review here the available literature on the reaction catalyzed by BSH enzymes, explore the ecological significance of BSH production, and briefly examine the impact that bile hydrolysis may have on human physiology. We conclude with suggestions for future work and possible applications of BSH research.
BILE
Bile is a yellow-green aqueous solution whose major constituents include bile acids, cholesterol, phospholipids, and the pigment biliverdin (12, 44). It is synthesized in the pericentral hepatocytes of the liver, stored and concentrated in the gallbladder interdigestively, and released into the duodenum after food intake. Bile functions as a biological detergent that emulsifies and solubilizes lipids, thereby playing an essential role in fat digestion. This detergent property of bile also confers potent antimicrobial activity, primarily through the dissolution of bacterial membranes (reviewed in reference 5).
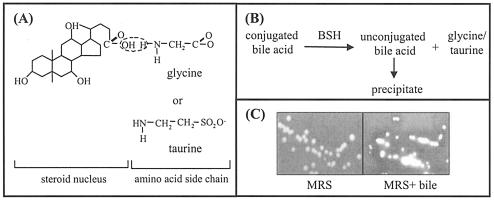
The primary bile acids, cholic and chenodeoxycholic acid, are synthesized de novo in the liver from cholesterol. The solubility of the hydrophobic steroid nucleus is increased by conjugation as an N-acyl amidate with either glycine (glycoconjugated) or taurine (tauroconjugated) prior to secretion (Fig. 1A). The resulting molecules are therefore amphipathic and can solubilize lipids to form mixed micelles.
FIG. 1.
(A) Chemical structure of bile acids. Primary bile acids are synthesized in the liver from cholesterol and are conjugated with either glycine or taurine prior to secretion. The carboxyl group of the bile acid and the amino group of the amino acid are linked by an amide bond. (B) Reaction catalyzed by BSH enzymes. BSHs cleave the peptide linkage of bile acids, which results in removal of the amino acid group from the steroid core. The resulting unconjugated bile acids precipitate at low pH. (C) Detection of BSH activity (as described in reference 18). L. plantarum, which was grown overnight in MRS broth, was streaked onto MRS (Difco) agar (A) or MRS agar supplemented with 0.2% (wt/vol) glycodeoxycholic acid (GDCA; Sigma) (B) and incubated anaerobically for 48 h. The white precipitates around colonies and the clearing of the medium are indicative of BSH activity (see reference 18).
Bile acids are efficiently conserved under normal conditions by a process termed enterohepatic recirculation. Conjugated and unconjugated bile acids are absorbed by passive diffusion along the entire gut and by active transport in the terminal ileum (12). Reabsorbed bile acids enter the portal bloodstream and are taken up by hepatocytes, reconjugated, and resecreted into bile. Approximately 5% of the total bile acid pool (0.3 to 0.6 g) per day eludes epithelial absorption and may be extensively modified by the indigenous intestinal bacteria (reviewed in reference 10). One important transformation is deconjugation, a reaction that must occur before further modifications are possible (3). Deconjugation is catalyzed by bile salt hydrolase (BSH) enzymes (EC 3.5.1.24), which hydrolyze the amide bond and liberate the glycine/taurine moiety from the steroid core (Fig. 1B). The resulting acids are termed unconjugated or deconjugated bile acids.
INCIDENCE OF BSH ACTIVITY AMONG BACTERIA
BSH activity has been detected in Lactobacillus (2, 13, 18, 21, 23, 30, 31, 39, 60, 61, 66, 72, 88, 90, 91), Bifidobacterium (40, 41, 50, 51, 52, 89), Enterococcus (34, 55, 106), Clostridium (14, 72), and Bacteroides (48, 85) spp. Lactobacilli and bifidobacteria are routinely used as probiotic strains, while Bacteroides, Clostridium, and Enterococcus spp. are also commensal inhabitants of the gastrointestinal tract. To date, BSH activity has not been detected in bacteria isolated from environments from which bile salts are absent (e.g., Lactococcus lactis or Streptococcus thermophilus) (1, 31, 68). With the exception of two strains of Bacteroides, all other BSH-positive bacteria are gram positive. All other gram-negative intestinal bacteria that have been examined (including Escherichia coli and Salmonella enterica serovar Typhimurium) neither demonstrate BSH activity nor possess bsh homologs in their genomes (M. Begley, unpublished data).
BSH in pathogenic bacteria.
The gram-positive gastrointestinal pathogen Listeria monocytogenes is not normally considered a member of the normal intestinal microbiota, but it does possess a BSH enzyme (Lmo2067 [6, 27]). However, since it is estimated that between 1 and 10% of the population asymptomatically shed L. monocytogenes, it has been suggested that this bacterium may be at the border between pathogenic and commensal microorganisms (27). Enterococcus faecalis, which is a normal inhabitant of the human gastrointestinal tract but can be an opportunistic pathogen, possesses a bsh homolog (EF0040; AAM75246) located within a pathogenicity island (83); however, this locus has not yet been functionally characterized. Dean et al. (20) purified a protein with bile-hydrolyzing activity from Xanthomonas maltophilia, a gram-negative opportunistic nosocomial pathogen that causes a wide variety of diseases but is most commonly recovered from the respiratory tract. However, N-terminal sequencing revealed that the protein was not similar to any characterized BSH. It is possible that it may not be a “true” BSH or, alternatively, the lack of homology may indicate a large divergence in the BSH family of proteins (the Ntn hydrolase family [discussed below]).
Identification of bsh homologs in probiotic genomes.
We looked for genes that may encode BSH enzymes in the genome sequences of potential probiotic bacteria available in public databases (using the National Center for Biotechnology Information genome site [http://www.ncbi.nlm.nih.gov/] and the Joint Genome Institute microbial genomics site [http://genome.jgi-psf.org/]). Consistent with previous findings, homologs were identified in all strains associated with the intestinal tract (Table 1). Interestingly, several strains (e.g., Lactobacillus plantarum WCFS1) possess more than one BSH homolog, which are not identical (data not shown). The potential significance of multiple BSH homologs is discussed below. Sequence analyses also revealed that the genetic geography of bsh regions is not the same in all strains, and in cases where more than one is present they are not located in the same region of the chromosome (data not shown).
TABLE 1.
BSH homologs in the genomes of sequenced probiotic strainsa
| Strainb | bsh homolog(s) (accession no.) | Size (aa) of predicted productc |
|---|---|---|
| Lactobacillus plantarum WCFS1* | lp_0067 (bsh 2) CAD62757 NP_783921 | 338 |
| lp_2572 (bsh 4) CAD64848 NP_785997 | 317 | |
| lp_3362 (bsh 3) CAD65471 NP_786598 | 328 | |
| lp_3536 (bsh 1) CAD65617 NP_786739QO6115 | 324 | |
| Lactobacillus johnsonii NCC533* | LJ1412 (NP_965212AAS09178) | 326 |
| LJ1147 (NP_965003AAS08969) | 325 | |
| LJ0056 (NP_964072AAS08038) | 316 | |
| Bifidobacterium longum NCC2705* | BLO796 (AAN24611NP_695975) | 317 |
| Lactobacillus acidophilus NCFM ATCC700396* | LBA1078 (bshB) (AAV42923YP_193954) | 325 |
| LBA0892 (AAV42751YP_193782) | 325 | |
| Lactobacillus brevis ATCC 367† | Scaffold 6, gene 1422 | 327 |
| Scaffold 29, gene 944 | 255 | |
| Lactobacillus gasseri ATCC 33323† | Scaffold 1, gene 149 | 325 |
| Scaffold 6, gene 1551 | 316 | |
| Bifidobacterium longum DJO10A† | Scaffold 1, revised gene 87 | 317 |
The information was collated in July 2005.
*, data were obtained from the National Center for Biotechnology Information genome site (http://www.ncbi.nlm.nih.gov/); †, data were obtained from the Joint Genome Institute microbial genomics site (http://genome.jgi-psf.org/).
aa, amino acids.
Horizontal transfer of bsh genes.
Since variability in BSH phenotypes has been observed within isolates of some species (15, 16, 31, 34, 88), it has been speculated that bsh genes may have been acquired horizontally (31). Comparison of the bsh gene and surrounding sequences of L. acidophilus strain KS-13 and L. johnsonii 100-100 by Elkins et al. (31) revealed little or no synteny flanking this locus. It was also noted that L. johnsonii 100-100 encodes a group II intron protein (maturase mat) downstream of bsh (31). In addition to reverse transcriptase activity, these proteins can function as maturases and endonucleases and facilitate movement and splicing of cDNA into the genome. Group II intron proteins are often inserted in or associated with mobile genetic elements (29). Sequencing of the entire genome of L. acidophilus NCFM revealed that this strain possesses two bsh genes (bshA and bshB). The predicted sequence of the BSH enzymes encoded by these loci share a higher level of similarity to BSH enzymes from other Lactobacillus species than to each other, suggesting that they may have been acquired from different sources (66).
The L. monocytogenes bsh gene is absent from the genome of the nonpathogenic strain L. innocua, whereas flanking regions have the same organization in both (27). In addition, the G+C content of the gene is lower than that of neighboring genes (36% versus 38 to 41%) but similar to the G+C content of the L. plantarum bsh, and the encoded protein shows most homology to the BSHs of lactobacilli (67% identity to the BSH of L. plantarum). These observations, together with the fact that L. monocytogenes shares the same microenvironments as lactobacilli during its life cycle (intestine, decaying vegetation, food, and vegetables) strongly suggest that the listerial gene may have been acquired from lactobacilli.
It has also been noted that the bsh gene in Bifidobacterium longum SBT2928 is flanked by inverted repeats (89), which may have played a role in the horizontal transfer of the gene (31). However, a study by Kim et al. (50) revealed that bsh genes were highly conserved within all bifidobacterial strains tested, and their G+C content reflected the overall G+C content of the genome. Also, since there are no reports of BSH activity from any other G+C-rich gram-positive bacteria and BSH activity has been observed among all bifidobacteria strains, it is likely that the bsh gene is a paralogous gene in the bifidobacterial genome (50).
In conclusion, BSH is present in all bifidobacterial strains and lactobacilli strains associated with the gastrointestinal environment, but bsh genes can potentially be acquired from these strains by other intestinal microorganisms (e.g., L. monocytogenes).
CHARACTERISTICS OF BSH ENZYMES
BSHs have been purified and characterized from various microorganisms (summarized in reference 49) and have generally found to be located intracellularly, are oxygen insensitive, and have slightly acidic pH optima (usually between pH 5 and 6) (13, 15, 16, 23, 36, 38, 41, 42, 48, 60, 85, 89, 93).
Active site.
BSHs belong to the choloylglycine hydrolase family of enzymes that also contains penicillin amidases (EC 3.5.1.11), enzymes that hydrolyze penicillin to yield 6-aminopenicillinic acid (6-APA). This intermediate is widely used in the industrial production of semisynthetic antibiotics (102). Both BSHs and penicillin amidases have been classified as N-terminal nucleophilic (Ntn) hydrolases with an N-terminal cysteine residue. This Cys-1 becomes a catalytic center after removal of the initiation formyl methionine by an autoproteolytic process, which is one of the common features of the Ntn hydrolase superfamily (86). The thiol (SH) group of Cys-1 has been shown to be essential for BSH catalysis. Replacement of the Bifidobacterium bifidum BSH Cys-1 with the nucleophilic amino acids serine or threonine which have a hydroxyl group instead of a thiol group abolishes BSH activity (50), and exchange of Cys-1 with alanine in the B. longum BSH results in an inactive protein (89). Agents that oxidize thiol groups (e.g., p-mercuribenzoate, iodoacetamide, Hg2+, Cu2+, and Cd2+) have been shown to strongly inhibit BSH activity in Clostridium perfringens (38), Lactobacillus reuteri (93), and B. longum (41, 89).
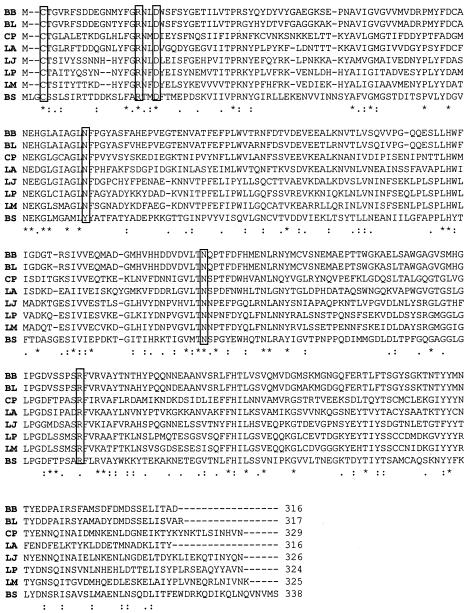
Other amino acids that are thought to play a role in BSH catalysis include Asp-20, Tyr-82, Asn-175, and Arg-228. Indeed, sequence alignments show that in addition to Cys-1 these amino acids are strictly conserved in all BSHs (Fig. 2). In addition, structural comparisons of the BSH of C. perfringens and other members of the Ntn hydrolase family have revealed that the geometry of the predicted active sites is well conserved (76).
FIG. 2.
CLUSTAL alignment of BSHs. Protein sequences were obtained from the National Center for Biotechnology Information site (http://www.ncbi.nlm.nih.gov/). Alignments were performed by using CLUSTAL W (http://www.ebi.ac.uk/clustalw/). Those amino acids thought to be involved in catalysis (Cys-1, Asp-21, Tyr-82, Asn-175, and Arg-228) are boxed. Identical amino acids are marked by an asterisk, conserved substitutions are marked by two dots, and semiconserved substitutions are marked by a single dot. BB, Bifidobacterium bifidum (AAT11513 [50]); BL, Bifidobacterium longum (AAF67801 [88]); CP, Clostridium perfringens (P54965 [14]); LA, Lactobacillus acidophilus (AAD03709 [31]); LJ, Lactobacillus johnsonii (AAG22541 [31]); LP, Lactobacillus plantarum (AAB24746 [13]); LM, Listeria monocytogenes (CAD00145 [5, 27]); and BS, Bacillus sphaericus (P12256 [70]).
Substrate specificity.
It is possible that BSHs recognize bile acids on both the cholate steroid nucleus and the amino acid groups (glycine/taurine). Recognition of the cholate group has been reported in the literature. A study by Moser and Savage (68) revealed that L. buchneri JCM1069 expressed taurodeoxycholic acid hydrolase activity but not taurocholic acid hydrolase activity. Taurodeoxycholic acid and taurocholic acid both have taurine as their amino acid moiety but differ at the 7α position of their steroid moieties. In addition, inactivation of bshA of L. acidophilus NCFM reduces the strain's ability to hydrolyze bile salts containing chenodeoxycholic as the steroid moiety, e.g., TCDCA and GCDCA (66). However, the majority of kinetic data available in the literature suggests that substrates are predominantly recognized at the amino acid moieties, and most BSHs are more efficient at hydrolyzing glycoconjugated bile salts than tauroconjugated bile salts (14, 51, 89, 93). Sequence alignments reveal that, whereas residues of the active site are strictly conserved in BSHs, the residues for substrate recognition are not particularly conserved although most amino acid substitutions are conservative (76). The notable exception is Leu142, which is strictly conserved, but the significance of this is unknown (56, 76).
Future structure analyses of BSHs from various species will undoubtedly discover key residues of the active site and substrate binding pocket and provide information on the substrate selectivity of BSH enzymes (56, 76). Such studies will also allow the comparison of the mechanism of action and the specificity of BSHs and penicillin amidases.
FUNCTION(S) OF BSH
The precise function(s) of microbial BSHs is currently unknown, although several hypotheses have been proposed (these are summarized in Table 2) and are discussed in this section.
TABLE 2.
Microbial role of BSH activity and impact of activity on the hosta
| Role or impact of BSH activity | References |
|---|---|
| Microbial role | |
| Bile detoxification | 1, 6, 23, 37, 39, 68, 101 |
| Gastrointestinal persistence | 2, 6, 27 |
| Nutritional role | 36, 45, 90, 103 |
| Membrane alterations (may increase resistance to bile, intestinal defensins, lysozyme, etc.) | 9, 17, 92, 94 |
| Impact on the host | |
| Altered digestive functions (lipid malabsorption, weight loss) | 22, 32, 33 |
| Cholesterol lowering | 22, 23, 28, 59, 73, 90, 95 |
| Cancer/activation of carcinogens | 7, 47, 65, 69, 71 |
| Formation of gallstones | 8, 62-64, 104 |
See the text for further details.
Nutritional role.
The amino acids liberated from bile salt deconjugation could potentially be used as carbon, nitrogen, and energy sources, since glycine may be metabolized to ammonia and carbon dioxide, and taurine may be metabolized to ammonia, carbon dioxide, and sulfate. Bile salt deconjugation may therefore confer a nutritional advantage on hydrolytic strains. In support of this hypothesis, Huijghebaert et al. (45) and Van Eldere et al. (103) observed that certain BSH-positive strains of Clostridium utilized the released taurine as an electron acceptor and that growth rates improved in the presence of taurine and taurine-conjugated bile salts. It has also been noted that transcription of the B. longum bsh gene is coupled to a homolog of glnE that encodes a glutamine synthetase adenyltransferase that forms part of the nitrogen regulation cascade (89). However, experiments performed by Tannock et al. (90) and Gilliland and Speck (36) refute this hypothesis since these authors observed that the lactobacilli used in their studies did not utilize the steroid moiety of the bile salt for cellular precursors since neither ring cleavage nor subsequent metabolism occurred.
Sequence analyses have revealed that BSHs show extensive homologies to penicillin amidases. Although they are commonly used in the industrial production of antibiotics, the microbial role of penicillin amidases is still not clear. Since they can cleave other phenyl and phenoxyacetic acid derivatives in addition to penicillins and are induced or repressed by different carbon sources, it has been suggested that penicillin amidases are involved in the assimilation of phenyl and phenoxyacetylated compounds (102). These compounds, such as phenolic and caffeic acids and flavonoids, are abundant in the environment since they are produced by the degradation of plant cell material by microorganisms. It is possible that BSHs may share the same substrates as penicillin amidases and, in addition to hydrolyzing bile, may also hydrolyze phenylacetylated compounds, thereby serving as scavengers in carbon-limiting conditions.
Alteration of membrane characteristics.
The bacteriolytic enzymes lysozyme and phospholipase A2, and antimicrobial peptides such as α-defensins, are important contributors to innate immunity in the intestine. The composition, fluidity, permeability, hydrophobicity, and net charge of bacterial membranes all determine the extent of damage by these host defenses (reviewed in reference 74). It has been proposed that BSHs facilitate incorporation of cholesterol or bile into bacterial membranes (17, 92, 94). This incorporation may increase the tensile strength of the membranes (9) or may change their fluidity or charge. Cell surface modifications that may result from BSH activity could potentially offer protection against perturbation of the structure and integrity of bacterial membranes by the immune system, and such resistance mechanisms may be important in establishing persistent infections. Such a function may strongly select for commensals possessing BSH enzymes while mitigating against BSH-negative pathogens or other transients.
Bile detoxification.
Studies by four independent groups using wild-type and bsh mutant pairs provide a link between bile salt hydrolysis and bile tolerance. A Lactobacillus amylovorus mutant with a partial decrease in BSH activity isolated using an N-methyl-N 1-nitro-N-nitrosoguanidine mutagenesis strategy displayed decreased growth rates in the presence of bile salts (39). Also, mutation of bsh in L. plantarum (23) and L. monocytogenes (6, 27) renders cells significantly more sensitive to bile and bile salts. The precise mechanism by which BSH enzymes play a role in the tolerance of bile is not yet fully understood. However, it has been proposed that since the protonated (nondissociated) form of bile salts may exhibit toxicity through intracellular acidification in a manner similar to organic acids, BSH-positive cells may protect themselves through the formation of the weaker unconjugated counterparts (23). This could help negate the drop in pH by recapturing and exporting the cotransported proton (23).
The ratio of glycoconjugated to tauroconjugated bile salts in human bile is usually 3:1 (44). In vitro experiments have revealed that whereas tauroconjugated bile salts usually only have slight affects (if any) on bacterial cells at every pH examined, glycoconjugated bile salts are extremely toxic at acidic pHs and bsh mutants are significantly more inhibited than corresponding parent cells (6, 23). We therefore suggest that BSHs are particularly important in combating the toxic effects of glycoconjugated bile salts at low pH, and BSH activity may be of particular importance at the point where bile enters the duodenum and where acid reflux may occur from the stomach, or in localized microenvironments in the intestine when the pH is lowered by lactic acid bacteria. The fact that BSHs have been shown to preferentially hydrolyze glycoconjugated bile salts (13, 14, 50, 89), together with the observation that BSHs have slightly acidic pH optima (usually between pH 5 and 6) (13, 15, 38, 41, 48, 59, 93), may serve to substantiate this theory.
Although several reports in the literature do not correlate the bile tolerance of strains with BSH activity (1, 37, 68, 92, 101), it is possible that these studies may have used inappropriate experimental conditions; for example, the use of tauroconjugated bile acids to detect BSH activity, even though the majority of BSHs show a preference for glycoconjugated bile acids. Furthermore, since many factors influence the bile tolerance of strains (e.g., membrane characteristics [reviewed in reference 5]), comparing nonisogenic strains will not give a true representation of the contribution of BSH to bile tolerance.
The unconjugated bile acids resulting from bile salt hydrolysis have greater inhibitory effects on bacteria than conjugated bile acids in vitro. However, it is possible that in vivo unconjugated products are precipitated at the low pHs in the intestine caused by fermentations by lactic acid bacteria. In fact, this localized precipitation phenomenon is the basis for the agar plate assay used to detect BSH activity (Fig. 1C) (18). It is also possible that BSH-active strains may be capable of detoxifying unconjugated bile acids as suggested by the study of De Smet et al. (23) or, alternatively, that they may associate with 7α-dehydroxylating bacteria that would dehydroxylate unconjugated bile acids (21).
Gastrointestinal persistence.
Since BSHs may combat the deleterious effects of bile (and perhaps components of the innate immune system such as the defensins through cell surface modifications), a role for these enzymes in survival/persistence of strains within the gastrointestinal tract is conceivable. Bateup et al. (2) compared the abilities of three Lactobacillus strains which demonstrated various degrees of BSH activity in vitro (one strain demonstrated high activity, one showed moderate activity, and one lacked activity) to colonize lactobacillus-free BALB/c mice. Enumeration of lactobacilli in the gastrointestinal organs 2 weeks after inoculation revealed that all strains colonized equally well, leading to the conclusion that BSH is not essential for colonization. However, a more recent study by Dussurget et al. (27) convincingly demonstrates that BSH contributes to persistence of L. monocytogenes within the gastrointestinal tract. A bsh mutant demonstrated reduced bacterial fecal carriage after oral infection of guinea pigs (counts of the mutant were 4 to 5 logs lower than the parent after 48 h). It was also observed that intestinal multiplication of the parent could be increased ∼10-fold by supplying cells with an extra copy of the gene on a plasmid, further confirming the importance of BSH to intestinal persistence (27). Two obvious differences between this L. monocytogenes study and the earlier one of Bateup et al. (2) may account for their different conclusions. First, isogenic L. monocytogenes wild-type and bsh mutant strains were compared, and it is possible that intrinsic differences between the strains of lactobacilli used in the other study masked the contribution of BSH to intestinal survival. Furthermore, Bateup et al. used Lactobacillus-free mice, and it is possible that a role for BSH would be uncovered in a more competitive environment. Future investigations with bifidobacterial and lactobacillus bsh mutants are necessary to unequivocally determine whether gastrointestinal persistence is a universal function of BSHs.
Multiple BSH homologs.
As mentioned previously, analyses of sequenced probiotic strains reveal that many possess more than one BSH homolog. The economic nature of bacterial genomes implies that possession of multiple BSHs should confer some advantage on strains. Each BSH may respond to different types of bile or perhaps different lengths of exposure to bile (e.g., bile adaptation), thereby ensuring maximal survival of the bacterium under changing environmental conditions. Recent experiments performed in both L. plantarum WCFS1, which has four bsh genes, and L. acidophilus NCFM, which has two bsh genes, supports this speculation (11, 66). In the study by Bron et al. (11), clone-based DNA microarrays were used to compare the transcriptional responses of cultures of L. plantarum WCFS1 that were grown for 3 days on de Man-Rogosa-Sharpe (MRS) agar containing 0.1% porcine bile to cultures grown in the absence of bile. Experiments revealed that bsh 1 (lp_3536) was induced ∼6-fold by the bile, while expression of bsh 3 (lp_3362) was reduced by ∼5-fold. Inactivation of the two bsh genes of L. acidophilus NCFM indicate that the encoded enzymes possess different substrate specificities. BSH A activity seems to be dictated by the steroid nucleus of bile salts, while the activity of BSH B is dictated by the amino acid side chain (66).
It is also possible that a strain possesses more than one BSH to fully take advantage of the other possible functions of these enzymes; for example, hydrolyzing bile to use as an energy source or perhaps incorporating it into the membrane to increase defense to membrane-damaging agents (e.g., bile or defensins). This may particularly be the fate of tauroconjugated bile acids, which do not seem to be very toxic to bacteria in a laboratory setting (4, 23). BSHs may also be capable of hydrolyzing phenylacetylated compounds either within the intestine or outside the host (e.g., in decaying vegetation or silage).
Targeted mutations in strains with multiple bsh homologs will permit detailed investigations into their precise functions. Enzyme assays and transcriptional analyses can be used to investigate the specificity of each enzyme toward different types of bile and bile salts, and persistence experiments will evaluate their role in intestinal survival.
IMPACT OF MICROBIAL BSH ACTIVITY ON THE HOST
Cholesterol lowering.
Hypercholesterolemia (elevated blood cholesterol levels) is considered a major risk factor for the development of coronary heart disease, and although pharmacologic agents are available to treat this condition (e.g., statins or bile acid sequestrants), they are often suboptimal and expensive and can have unwanted side effects (81). Oral administration of probiotics has been shown to significantly reduce cholesterol levels by as much as 22 to 33% (22, 24, 28, 90, 95; reviewed in reference 73) or prevent elevated cholesterol levels in mice fed a fat-enriched diet (96). These cholesterol-lowering effects can be partially ascribed to BSH activity (other possible mechanisms not discussed here include assimilation of cholesterol by the bacteria, binding of cholesterol to the bacterial cell walls, or physiological actions of the end products of short-chain fatty acid fermentation) (54, 59). Deconjugated bile salts are less efficiently reabsorbed than their conjugated counterparts, which results in the excretion of larger amounts of free bile acids in feces. Also, free bile salts are less efficient in the solubilization and absorption of lipids in the gut. Therefore, deconjugation of bile salts could lead to a reduction in serum cholesterol either by increasing the demand for cholesterol for de novo synthesis of bile acids to replace those lost in feces or by reducing cholesterol solubility and thereby absorption of cholesterol through the intestinal lumen.
Impaired digestive functions.
Since unconjugated bile acids are less efficient than conjugated molecules in the emulsification of dietary lipids and the formation of micelles, BSH activity may compromise normal lipid digestion, and the absorption of fatty acids and monoglycerides could be impaired (22). Microbial BSH activity has been related to growth defects in chickens (32, 33) but not in mice (2).
Disruption of normal intestinal conditions and/or gallstones.
It has been proposed that secondary bile acids resulting from the subsequent modification of unconjugated bile salts may cause DNA damage, promote colon cancer, or result in impaired colonic mucosal function that would lead to diarrhea or inflammation (7, 47, 65, 69, 71). In addition, since the solubilization of cholesterol in bile depends on the ratio of cholesterol to bile salts and lecithin, alterations in the concentrations of bile acids may result in bile being supersaturated with cholesterol. This cholesterol may precipitate together with calcium salts and bile pigments to form concretions termed gallstones, which may grow and obstruct the biliary ducts (62, 64, 104). It is noteworthy that an increase in secondary bile acids has been observed in gallstone sufferers (8, 63).
IS BSH ACTIVITY A DESIRABLE TRAIT IN PROBIOTICS?
Overall, the data strongly support the hypothesis that microbial BSHs function in the detoxification of bile salts and in doing so increase the intestinal survival and persistence of producing strains. Therefore, BSH activity by a probiotic bacterium may be desirable since it could maximize its prospects of survival in the hostile environment of the gastrointestinal tract. Increased intestinal survival is likely to increase the overall beneficial effects associated with the strain.
Since large amounts of deconjugated bile salts may have undesirable effects for the human host (described earlier), concerns may arise over the safety of administering a BSH-positive probiotic strain. However, the bacterial genera that would most likely be used as probiotics (bifidobacteria and lactobacilli) are not capable of dehydroxylating deconjugated bile salts (1, 36, 87), and so the majority of the breakdown products of BSH activity by a probiotic strain may be precipitated and excreted in feces (this may vary from person to person depending on colonic pH and intestinal transit time [97, 98, 105]). In addition, work performed by two groups has shown that it may be possible to prevent further modification of deconjugated products by other intestinal microorganisms (e.g., certain strains of Clostridium and Eubacterium, which are the only strains that have been shown to possess dehydroxylating activity [19, 25]). First, Jones et al. (46) investigated the ability of a BSH-positive L. plantarum strain encapsulated in an artificial membrane to hydrolyze bile salts. The experiments of Jones et al. demonstrated that the microencapsulated strain was able to effectively break down physiologically relevant concentrations of bile in vitro, but the products of BSH deconjugation were trapped within the membrane. In addition to increasing the safety of the strain by rendering these products less bioavailable, microencapsulation would also protect entrapped bacteria from harsh environmental conditions encountered during gastric transit. Second, studies by Kurdi et al. (57, 58) revealed that cholic acid, the main free bile acid produced by BSH activity in the intestine, could accumulate inside the bifidobacterium and lactobacillus strains examined so long as the bacteria were energized. The amount of accumulation increased at decreasing external pH values, suggesting that factors which decrease the intestinal pH (the presence of short-chain fatty acids or lactic acid produced by intestinal microbes) may enhance the accumulation of cholic acid by lactobacilli in vivo (57).
In summary, BSH activity may benefit a probiotic bacterium required to survive and perform in the intestinal milieu. Microencapsulating the bacterium or selection of a strain that is not capable of further modifying unconjugated bile salts or one that may accumulate them would address the medical concerns about the possible side effects associated with BSH activity.
FUTURE DIRECTIONS AND APPLICATIONS OF BSH RESEARCH
Directions for future studies.
Future investigations should concentrate on ascertaining the precise role of BSH enzymes in gastrointestinal bacteria. Studies to elucidate the fate of the end products of bile hydrolysis and to compare the behavior and characteristics of targeted bsh mutants to their isogenic parent strains are of vital importance. Examination of the membrane properties (charge, hydrophobicity, fatty acid composition, etc.) of these strains grown in the absence and presence of bile would also be revealing. Furthermore, controlled animal trials will help to uncover the contribution of BSH to intestinal survival. From the host perspective molecular tools such as denaturing gradient gel electrophoresis and fluorescence in situ hybridization can be used to monitor the impact of administering a BSH-positive strain on the established gut microflora. Also, since secondary bile acid production has been considered as a significant risk factor for the development of colon cancer, it must be determined whether serum or biliary levels of secondary bile acids increase in the bile acid pool as a result of bile salt hydrolysis.
Possible applications of BSH research.
A better understanding of the role of BSH may be exploited in the selection and rational design of probiotic strains. Since it is likely that BSH significantly contributes to the bile tolerance and survival and persistence of strains in the intestinal tract, it may be desirable to select probiotic bacteria that possess these enzymes. Banks of strains could be screened for the presence of bsh genes by performing PCRs with degenerate primers based on conserved regions of BSH enzymes, BSH activity can then be examined by using the well-established agar plate assay (18). Such an approach may allow a rapid and simple screen of banks for strains capable of performing in the intestinal tract a task that is currently tedious and time-consuming. It may also be possible to manipulate the BSH activity of probiotic strains (either to overexpress a native BSH or to express or overexpress a heterologous BSH) to improve their survivability in the intestinal tract. Finally, administration of a bile-hydrolyzing strain to control serum cholesterol levels (i.e., oral live bacterial cell therapy) shows much promise. This “biological approach” may especially be appealing to an emerging health conscious society since the ingestion of a probiotic-containing food may be considered more “natural” than other cholesterol-lowering therapies.
Conclusion.
It is becoming increasingly obvious that BSH enzymes may confer a selective advantage on probiotic strains in the highly competitive environment of the human intestinal tract, and future investigations will reveal the exact extent of their contribution(s). Manipulation of BSH activity may ultimately lead to the development of more robust probiotics with improved competitiveness and performance.
Acknowledgments
We acknowledge funding received from the Irish Government under the National Development Plan 2000-2006 and from the Alimentary Pharmabiotic Centre by the Science Foundation of Ireland Centres for Science Engineering and Technology (CSET) scheme.
REFERENCES
- 1.Ahn, Y. T., G. B. Kim, Y. S. Lim, Y. J. Baek, and Y. U. Kim. 2003. Deconjugation of bile salts by Lactobacillus acidophilus isolates. Int. Dairy J. 13:303-311. [Google Scholar]
- 2.Bateup, J. M., M. A. McConnell, H. F. Jenkinson, and G. W. Tannock. 1995. Comparison of Lactobacillus strains with respect to bile salt hydrolase activity, colonization of the gastrointestinal tract, and growth rate of the murine host. Appl. Environ. Microbiol. 61:1147-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batta, A. K., G. Salen, R. Arora, S. Shefer, M. Batta, and A. Perseon. 1990. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J. Biol. Chem. 265:10925-10928. [PubMed] [Google Scholar]
- 4.Begley, M., C. G. M. Gahan, and C. Hill. 2002. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl. Environ. Microbiol. 68:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begley, M., C. G. M. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29:625-651. [DOI] [PubMed] [Google Scholar]
- 6.Begley, M., R. D. Sleator, C. G. M. Gahan, and C. Hill. 2005. The contribution of three bile-associated loci (bsh, pva, and btlB) to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73:894-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, C., H. Bernstein, C. M. Payne, K. Dvorakova, and H. Garewal. 2005. Bile acids as carcinogens in human gastrointestinal cancers. Mutat. Res. 589:47-65. [DOI] [PubMed] [Google Scholar]
- 8.Berr, F., G. A. Kullak-Ublick, G. Paumgartner, W. Munzing, and P. B. Hylemon. 1996. 7α-Dehydroxylating bacteria enhance deoxycholic acid input and cholesterol saturation of bile in patients with gallstones. Gastroenterology 111:1611-1620. [DOI] [PubMed] [Google Scholar]
- 9.Boggs, J. M. 1987. Lipid intermolecular hydrogen bonding: influence on structural organization and membrane function. Biochim. Biophys. Acta 906:353-404. [DOI] [PubMed] [Google Scholar]
- 10.Bortolini, O., A. Medici, and S. Poli. 1997. Biotransformations on steroid nucleus of bile acids. Steroids 62:564-577. [DOI] [PubMed] [Google Scholar]
- 11.Bron, P. A., D. Molenaar, W. M. De Vos, and M. Kleerebezem. DNA micro-array based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 12.Carey, M. C., and W. C. Duane. 1994. Enterohepatic circulation, p. 719-738. In I. M. Arias, N. Boyer, N. Fausto, W. B. Jackoby, D. A. Schachter, and D. A. Shafritz (ed.), The liver: biology and pathobiology. Raven Press, Ltd., New York, N.Y.
- 13.Christiaens, H., R. J. Leer, P. H. Pouwels, and W. Verstraete. 1992. Cloning and expression of a conjugated bile acid hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl. Environ. Microbiol. 58:3792-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman, J. P., and L. L. Hudson. 1995. Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl. Environ. Microbiol. 61:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corzo, G., and S. E. Gilliland. 1999. Bile salt hydrolase activity of three strains of Lactobacillus acidophilus. J. Dairy Sci. 82:472-480. [DOI] [PubMed] [Google Scholar]
- 16.Corzo, G., and S. E. Gilliland. 1999. Measurement of bile salt hydrolase activity from Lactobacillus acidophilus based on disappearance of conjugated bile salts. J. Dairy Sci. 82:466-471. [DOI] [PubMed] [Google Scholar]
- 17.Dambekodi, P. C., and S. E. Gilliland. 1998. Incorporation of cholesterol into the cellular membrane of Bifidobacterium longum. J. Dairy Sci. 81:1818-1824. [DOI] [PubMed] [Google Scholar]
- 18.Dashkevicz, M. P., and S. D. Feighner. 1989. Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl. Environ. Microbiol. 55:11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson, J. A., D. H. Mallonee, I. Björkhem, and P. B. Hylemon. 1996. Expression and characterization of a C24 bile acid 7α-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J. Lipid Res. 37:1258-1267. [PubMed] [Google Scholar]
- 20.Dean, M., C. Cervellati, E. Casanova, M. Squerzanti, V. Lanzara, A. Medici, P. Polverino de Laureto, and C. M. Bergamini. 2002. Characterization of cholylglycine hydrolase from a bile-adapted strain of Xanthomonas maltophilia and its application for quantitative hydrolysis of conjugated bile salts. Appl. Environ. Microbiol. 68:3126-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Boever, P., and W. Verstraete. 1999. Bile salt deconjugation by Lactobacillus plantarum 80 and its implication for bacterial toxicity. J. Appl. Microbiol. 87:345-352. [DOI] [PubMed] [Google Scholar]
- 22.De Smet, I., I. Van Hoorde, M. De Saeyer, W. M. Vande, and W. Verstraete. 1994. In vitro study of bile salt hydrolase (bsh) activity of bsh isogenic Lactobacillus plantarum 80 strains and estimation of cholesterol lowering through enhanced BSH activity. Microbial Ecol. Health Dis. 7:315-329. [Google Scholar]
- 23.De Smet, I., L. Van Hoorde, M. Vande Woestyne, H. Christiaens, and W. Verstraete. 1995. Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 79:292-301. [DOI] [PubMed] [Google Scholar]
- 24.De Smet, I., P. De Boever, and W. Verstraete. 1998. Cholesterol lowering in pigs through enhanced bacterial bile salt hydrolase activity. Br. J. Nutr. 79:185-194. [DOI] [PubMed] [Google Scholar]
- 25.Doerner, K. C., F. Takamine, C. P. LaVoie, D. H. Mallonee, and P. B. Hylemon. 1997. Assessment of fecal bacteria with bile acid 7α-dehydroxylation activity for the presence of bai-like genes. Appl. Environ. Microbiol. 63:1185-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunne, C., L. O' Mahony, L. Murphy, G. Thorton, D. Morrissey, S. O' Halloran, M. Feeney, S. Flynn, G. Fitzgerald, C. Daly, B. Kiely, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 2001. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 73:386S-392S. [DOI] [PubMed] [Google Scholar]
- 27.Dussurget, O., D. Cabanes, P. Dehoux, M. Lecuit, C. Buchreiser, P. Glaser, P. Cossart, et al. 2002. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45:1095-1106. [DOI] [PubMed] [Google Scholar]
- 28.du Toit, M., C. M. Franz, L. M. Dicks, U. Schillinger, P. Harberer, B. Warlies, F. Ahrens, and W. H. Holzapfel. 1998. Characterisation and selection of probiotic lactobacilli for a preliminary minipig feeding trial and their effect on serum cholesterol levels, faeces pH, and faeces moisture content. Int. J. Food Microbiol. 40:93-104. [DOI] [PubMed] [Google Scholar]
- 29.Edgell, D. R., M. Belfort, and D. A. Shub. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elkins, E. A., and D. C. Savage. 1998. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J. Bacteriol. 180:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkins, C. A., S. A. Moser, and D. C. Savage. 2001. Genes encoding bile salt hydrolases and conjugated bile salt transporters in Lactobacillus johnsonii 100-100 and other Lactobacillus species. Microbiology 147:3403-3412. [DOI] [PubMed] [Google Scholar]
- 32.Feighner, S. D., and M. P. Dashkevicz. 1987. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl. Environ. Microbiol. 53:331-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feighner, S. D., and M. P. Dashkevicz. 1988. Effect of dietary carbohydrates on bacterial cholyltaurine hydrolase in poultry intestinal homogenates. Appl. Environ. Microbiol. 54:337-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franz, C. M. A. P., I. Specht, P. Haberer, and W. H. Holzapfel. 2001. Bile salt hydrolase activity of enterococci isolated from food: screening and quantitative determination. J. Food Prot. 64:725-729. [DOI] [PubMed] [Google Scholar]
- 35.Gill, H. S., and F. Guarner. 2004. Probiotics and human health: a clinical perspective. Postgrad. Med. J. 80:516-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilliland, S. E., and M. J. Speck. 1977. Deconjugation of bile acids by intestinal lactobacilli. Appl. Environ. Microbiol. 33:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gopal, A., N. P. Shah, and H. Roginski. 1996. Bile tolerance, taurocholate deconjugation, and cholesterol removal by Lactobacillus acidophilus and Bifidobacterium spp. Milchwissenschaft 51:619-623. [Google Scholar]
- 38.Gopal-Srivastava, R., and P. B. Hylemon. 1988. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 29:1079-1085. [PubMed] [Google Scholar]
- 39.Grill, J. P., C. Cayuela, J. M. Antoine, and F. Schneider. 2000. Isolation and characterization of a Lactobacillus amylovorus mutant depleted in conjugated bile salt hydrolase activity: relation between activity and bile salt resistance. J. Appl. Microbiol. 89:553-563. [DOI] [PubMed] [Google Scholar]
- 40.Grill, J. P., C. Maginot-Durr, F. Schneider, and J. Ballongue. 1995. Bifidobacteria and probiotic effects: action of Bifidobacterium species on conjugated bile salts. Curr. Microbiol. 31:23-27. [DOI] [PubMed] [Google Scholar]
- 41.Grill, J. P., F. Schneider, J. Crociani, and J. Ballongue. 1995. Purification and characterization of conjugated bile salt hydrolase from Bifidobacterium longum BB536. Appl. Environ. Microbiol. 61:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grill, J. P., S. Perrin, and F. Schneider. 2000. Bile toxicity to some bifidobacteria strains: role of conjugated bile salt hydrolase and pH. Can. J. Microbiol. 46:878-884. [DOI] [PubMed] [Google Scholar]
- 43.Guarner, F., and G. Schaafsma. 1998. Probiotics. Int. J. Food Microbiol. 39:237-238. [DOI] [PubMed] [Google Scholar]
- 44.Hofmann, A. F. 1994. Bile acids, p. 677-718. In I. M. Arias, J. L. Boyer, N. Fausto, W. B. Jackoby, D. A. Schachter, and D. A. Shafritz (ed.), The liver: biology and pathobiology. Raven Press, Ltd., New York, N.Y.
- 45.Huijghebaert, S. M., J. A. Mertens, and H. J. Eyssen. 1982. Isolation of a bile salt sulfatase-producing Clostridium strain from rat intestinal microflora. Appl. Environ. Microbiol. 43:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones, M. L., H. Chen, W. Ouyang, T. Metz, and S. Prakash. 2004. Microencapsulated genetically engineered Lactobacillus plantarum 80 (pCBH1) for bile acid deconjugation and its implication in lowering cholesterol. J. Biomed. Biotechnol. 1:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kandell, R. L., and C. Bernstein. 1991. Bile salt/acid induction of DNA damage in bacterial and mammalian cells: implications for colon cancer, Nutr. Cancer 16:227-238. [DOI] [PubMed] [Google Scholar]
- 48.Kawamoto, K., I. Horibe, and K. Uchida. 1989. Purification and characterization of a new hydrolase for conjugated bile acids, chenodeoxycholyltaurine hydrolase, from Bacteroides vulgatus. J. Biochem. 106:1049-1053. [DOI] [PubMed] [Google Scholar]
- 49.Kim, G. B., and B. H. Lee. 2005. Biochemical and molecular insights into bile salt hydrolase in the gastrointestinal microflora: a review. Asian-Aust. J. Anim. Sci. 18:1505-1512. [Google Scholar]
- 50.Kim, G. B., C. M. Miyamoto, E. A. Meighen, and B. H. Lee. 2004. Cloning and characterization of the bile salt hydrolase genes (bsh) from Bifidobacterium bifidium strains. Appl. Environ. Microbiol. 70:5603-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim, G. B., S. H. Yi, and B. H. Lee. 2004. Purification and characterisation of three different types of bile salt hydrolase from Bifidobacterium strains. J. Dairy Sci. 87:258-266. [DOI] [PubMed] [Google Scholar]
- 52.Kim, G. B., M. Brochet, and B. H. Lee. 2005. Cloning and characterization of a bile salt hydrolase (bsh) from Bifidobacterium adolescentis. Biotechnol. Lett. 27:817-822. [DOI] [PubMed] [Google Scholar]
- 53.Klaenhammer, T. R., and M. J. Kullen. 1999. Selection and design of probiotics. Int. J. Food Microbiol. 50:45-57. [DOI] [PubMed] [Google Scholar]
- 54.Klaver, F. A. M., and R. Van der Meer. 1993. The assumed assimilation of cholesterol by lactobacilli and Bifidobacterium bifidum is due to their bile salt-deconjugating activity. Appl. Environ. Microbiol. 59:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knarreborg, A., R. M. Engberg, S. K. Jensen, and B. B. Jensen. 2002. Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens. Appl. Environ. Microbiol. 68:6425-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar, R. S., J. A. Brannigan, A. Pundle, A. Prabhune, G. G. Dodson, and C. G. Suresh. 2004. Expression, purification, crystallization, and preliminary X-ray diffraction analysis of conjugated bile salt hydrolase from Bifidobacterium longum. Acta Crystallogr. D Biol. Crystallogr. 60:1665-1667. [DOI] [PubMed] [Google Scholar]
- 57.Kurdi, P., H. Tanaka, H. W. van Veen, K. Asano, F. Tomita, and A. Yokota. 2003. Cholic acid accumulation and its diminution by short-chain fatty acids in bifidobacteria. Microbiology 149:2031-2037. [DOI] [PubMed] [Google Scholar]
- 58.Kurdi, P., H. W. van Veen, H. Tanaka, I. Mierau, W. N. Konings, G. W. Tannock, F. Tomita, and A. Yokota. 2000. Cholic acid is accumulated spontaneously, driven by membrane ΔpH, in many lactobacilli. J. Bacteriol. 182:6525-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liong, M. T., and N. P. Shah. 2005. Bile salt deconjugation ability, bile salt hydrolase activity and cholesterol co-precipitation ability of lactobacilli strains. Int. Dairy J. 15:391-398. [Google Scholar]
- 60.Lundeen, S. G., and D. C. Savage. 1990. Characterization and purification of bile salt hydrolase from Lactobacillus sp. strain 100-100. J. Bacteriol. 172:4171-4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lundeen, S. G., and D. C. Savage. 1992. Multiple forms of bile salt hydrolase from Lactobacillus sp. strain 100-100. J. Bacteriol. 174:7217-7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Low-Beer, T. S., and S. Nutter. 1978. Colonic bacterial activity, biliary cholesterol saturation, and pathogenesis of gallstones. Lancet ii:1063-1065. [DOI] [PubMed] [Google Scholar]
- 63.Mamianett, A., D. Garrido, C. N. Carducci, and M. C. Vescina. 1999. Fecal bile acid excretion pattern profile in gallstone patients. Medicina 59:269-273. [PubMed] [Google Scholar]
- 64.Marcus, S. N., and K. W. Heaton. 1986. Intestinal transit, deoxycholic acid, and the cholesterol saturation of bile: three inter-related factors. Gut 27:550-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marteau, P., M. F. Gehard, A. Myara, E. Bouvier, F. Trivin, and J. C. Rambaud. 1995. Metabolism of bile salts by alimentary bacteria during transit in the human small intestine. Microb. Ecol. Health Dis. 8:151-157. [Google Scholar]
- 66.Mc Auliffe, O., R. J. Cano, and T. R. Klaenhammer. 2005. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71:4925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mercenier, A., S. Pavan, and B. Pot. 2003. Probiotics as biotherapeutic agents: present knowledge and future prospects. Curr. Pharm. Des. 8:99-110. [DOI] [PubMed] [Google Scholar]
- 68.Moser, S. A., and D. C. Savage. 2001. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 67:3476-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nagengast, F. M., M. J. Grobben, and I. P. Van Munster. 1995. Role of bile acids in colorectal carcinogenesis. Eur. J. Cancer 31:1067-1070. [DOI] [PubMed] [Google Scholar]
- 70.Olsson, A., and M. Uhlen. 1986. Sequencing and heterologous expression of the gene encoding penicillin V amidase from Bacillus sphaericus. Gene 45:175-181. [DOI] [PubMed] [Google Scholar]
- 71.Pazzi, P., A. C. Puriani, M. Dalla Libera, G. Guerra, D. Rici, S. Gullini, and C. Ottolenghi. 1997. Bile salt-induced cytotoxicity and ursodeoxycholate cytoprotection: in vitro study in perfused rat hepatocytes. Eur. J. Gastroenterol. Hepatol. 9:703-709. [DOI] [PubMed] [Google Scholar]
- 72.Pereira, D. I., A. L. McCartney, and G. R. Gibson. 2003. An in vitro study of the probiotic potential of a bile-salt-hydrolyzing Lactobacillus fermentum strain, and determination of its cholesterol-lowering properties. Appl. Environ. Microbiol. 69:4743-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pereira, D. I., and G. R. Gibson. 2002. Effects of consumption of probiotics and prebiotics on serum lipid levels in humans. Crit. Rev. Biochem. Mol. Biol. 37:259-281. [DOI] [PubMed] [Google Scholar]
- 74.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 75.Reid, G., J. Jass, M. T. Sebulsky, and J. K. McCormick. 2003. Potential uses of probiotics in clinical practice. Clin. Microbiol. Rev. 16:658-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rossocha, M., R. Schultz-Heienbrok, H. von Moeller, J. P. Coleman, and W. Saenger. 2005. Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry 44:5739-5748. [DOI] [PubMed] [Google Scholar]
- 77.Saarela, M., G. Mogensen, R. Fondén, J. Mättö, and T. Mattila-Sandholm. 2000. Probiotic bacteria: safety, functional and technological properties. J. Biotechnol. 84:197-215. [DOI] [PubMed] [Google Scholar]
- 78.Sánchez, B., M. C. Champomier-Vergès, P. Anglade, F. Baraige, C. G. de los Reyes-Gavilán, A. Margolles, and M. Zagores. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savijoki, K., A. Suokko, A. Palva, L. Valmu, N. Kalkkinen, and P. Varmanen. 2005. Effect of heat-shock and bile salts on protein synthesis of Bifidobacterium longum revealed by [35S]methionine labeling and two-dimensional gel electrophoresis. FEMS Microbiol. Lett. 248:207-215. [DOI] [PubMed] [Google Scholar]
- 80.Sanders, M. E. 1999. Probiotics. Food Technol. 53:67-77. [Google Scholar]
- 81.Schuster, H. 2004. Improving lipid management: to titrate, combine, or switch. Int. J. Clin. Pract. 58:689-694. [DOI] [PubMed] [Google Scholar]
- 82.Shanahan, F. 2003. Probiotics in inflammatory bowel disease: therapeutic rationale and role. Adv. Drug Deliv. Rev. 56:809-818. [DOI] [PubMed] [Google Scholar]
- 83.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 84.Stanton, C., G. Gardiner, H. Meehan, K. Collins, G. Fitzgerald, P. B. Lynch, and R. P. Ross. 2001. Market potential for probiotics. Am. J. Clin. Nutr. 73(Suppl.):476S-483S. [DOI] [PubMed] [Google Scholar]
- 85.Stellwag, E. J., and P. B. Hylemon. 1976. Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim. Biophys. Acta 452:165-176. [DOI] [PubMed] [Google Scholar]
- 86.Suresh, C. G., A. V. Pundle, H. SivaRaman, K. N. Rao, J. A. Brannigan, C. E. McVey, C. S. Verma, Z. Dauter, E. J. Dodson, and G. G. Dodson. 1999. Penicillin V acylase crystal structure reveals new Ntn-hydrolase family members. Nat. Struct. Biol. 6:414-416. [DOI] [PubMed] [Google Scholar]
- 87.Takahashi, T., and M. Morotomi. 1994. Absence of cholic acid 7α-dehydroxylase activity in the strains of Lactobacillus and Bifidobacterium. J. Dairy Sci. 77:3275-3286. [DOI] [PubMed] [Google Scholar]
- 88.Tanaka, H., K. Doesburg, T. Iwasaki, and I. Mierau. 1999. Screening of lactic acid bacteria for bile salt hydrolase activity. J. Dairy Sci. 82:2530-2535. [DOI] [PubMed] [Google Scholar]
- 89.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum: biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tannock, G. W., M. P. Dashkevicz, and S. D. Feighner. 1989. Lactobacilli and bile salt hydrolase in the murine intestinal tract. Appl. Environ. Microbiol. 55:1848-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Taranto, M. P., D. G. De Llano, A. Rodriguez, A. P. De Ruiz Holgado, and G. Font de Valdez. 1996. Bile tolerance and cholesterol reduction by Enterococcus faecium, a candidate microorganism for the use as a dietary adjunct in milk products. Milchwissenschaft 51:383-385. [Google Scholar]
- 92.Taranto, M. P., F. Sesma, A. P. Ruiz Holgado, and G. Font de Valdez. 1997. Bile salt hydrolase plays a key role on cholesterol removal by Lactobacillus reuteri. Biotechnol. Lett. 19:845-847. [Google Scholar]
- 93.Taranto, M. P., and G. Font de Valdez. 1999. Localization and primary characterization of bile salt hydrolase from Lactobacillus reuteri. Biotechnol. Lett. 21:935-938. [Google Scholar]
- 94.Taranto, M. P., M. L. Fernandez Murga, G. Lorca, and G. Font de Valdez. 2003. Bile salts and cholesterol induce changes in the lipid cell membrane of Lactobacillus reuteri. J. Appl. Microbiol. 95:86-91. [DOI] [PubMed] [Google Scholar]
- 95.Taranto, M. P., M. Medici, G. Perdigon, A. P. Ruiz Holgado, and G. F. Valdez. 1998. Evidence for hypocholesterolemic effect of Lactobacillus reuteri in hypercholesterolemic mice. J. Dairy Sci. 81:2336-2340. [DOI] [PubMed] [Google Scholar]
- 96.Taranto, M. P., M. Medici, G. Perdigon, A. P. Ruiz Holgado, and G. F. Valdez. 2000. Effect of Lactobacillus reuteri on the prevention of hypercholesterolemia in mice. J. Dairy Sci. 83:401-403. [DOI] [PubMed] [Google Scholar]
- 97.Thomas, L. A., M. J. Veysey, T. Bathgate, A. King, G. French, N. C. Smeeton, G. M. Murphy, and R. H. Dowling. 2000. Mechanism for the transit-induced increase in colonic deoxycholic acid formation in cholesterol cholelithiasis. Gastroenterology 119:806-815. [DOI] [PubMed] [Google Scholar]
- 98.Thomas, L. A., M. J. Veysey, G. M. Murphy, and R. H. Dowling. 2001. Influence of pH on the phase distribution of nascent deoxycholic acid in fresh human cecal aspirates. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G371-G374. [DOI] [PubMed] [Google Scholar]
- 99.Tuohy, K. M., H. M. Probert, C. W. Smejkal, and G. R. Gibson. 2003. Using probiotics and prebiotics to improve gut health. Drug Discov. Today 8:692-700. [DOI] [PubMed] [Google Scholar]
- 100.Tuomola, E., R. Crittenden, M. Playne, E. Isolauri, and S. Salminen. 2001. Quality assurance criteria for probiotic bacteria. Am. J. Clin. Nutr. 73:393S-398S. [DOI] [PubMed] [Google Scholar]
- 101.USMAN (The United Graduate School of Agricultural Science), and A. Hosono. 1999. Bile tolerance, taurocholate deconjugation, and binding of cholesterol by Lactobacillus gasseri strains. J. Dairy Sci. 82:243-248. [DOI] [PubMed] [Google Scholar]
- 102.Valle, F., P. Balbás, E. Merino, and F. Bolivar. 1991. The role of penicillin amidases in nature and industry. Trends Biochem. Sci. 16:36-40. [DOI] [PubMed] [Google Scholar]
- 103.Van Eldere, J., P. Celis, G. De Pauw, E. Lesaffre, and H. Eyssen. 1996. Tauroconjugation of cholic acid stimulates 7α-dehydroxylation by fecal bacteria. Appl. Environ. Microbiol. 62:656-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Veysey, M. J., L. A. Thomas, A. I. Mallet, P. J. Jenkins, G. M. Besser, J. A. Wass, G. M. Murphy, and R. H. Dowling. 1999. Prolonged large bowel transit increases serum deoxycholic acid: a risk factor for octreotide induced gallstones. Gut 44:675-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Veysey, M. J., L. A. Thomas, A. I. Mallet, P. J. Jenkins, G. M. Besser, G. M. Murphy, and R. H. Dowling. 2001. Colonic transit influences deoxycholic acid kinetics. Gastroenterology 121:812-822. [DOI] [PubMed] [Google Scholar]
- 106.Wijaya, A., A. Hermann, H. Abriouel, I. Specht, N. M. Yousif, W. H. Holzapfel, and C. M. Franz. 2004. Cloning of the bile salt hydrolase (bsh) gene from Enterococcus faecium FAIR-E-345 and chromosomal location of bsh genes in food enterococci. J. Food Prot. 67:2772-2778. [DOI] [PubMed] [Google Scholar]