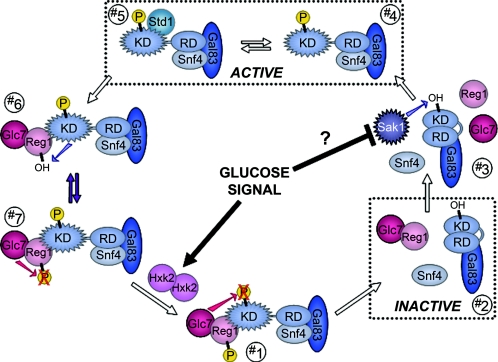
FIG. 3.
The opposing roles of Reg1/Glc7 phosphatase and the Snf1 kinase complex. The duel between Reg1/Glc7 and Snf1/Snf4/Gal83 turns the Snf1 kinase on (ACTIVE) and off (INACTIVE) by determining whether the Snf1 regulatory domain (RD) binds to and autoinhibits its kinase domain (KD). This switch plays a central role in determining the transcriptomic response to the presence or absence of glucose. The β subunit Gal83 acts as a scaffolding factor for the nuclear form of the Snf1 (α) and Snf4 (γ) subunits of the kinase. Addition of glucose to yeast cells growing on an alternative carbon source results in the dephosphorylation of active Snf1 kinase by Reg1/Glc7 (no. 1); this returns Snf1 to the autoinhibited state (no. 2) and results in export of Snf1/Gal83 to the cytoplasm (not shown). Once glucose is depleted, Sak1 phosphorylates Snf1 (no. 3), Snf1/Gal83 enters the nucleus (not shown), and Snf4 binds to the Snf1 regulatory domain, thereby releasing the catalytic domain (no. 4); Reg1 then undergoes rapid Snf1-dependent phosphorylation (no. 6), which stabilizes the interaction between Snf1 and the Reg1/Glc7 phosphatase (no. 7) and primes the latter to repress Snf1 anew should glucose reappear. Std1 interacts with the Snf1 catalytic domain and stimulates kinase activity (no. 5), although it is stoichiometrically underrepresented in Snf1 complexes. A dynamic equilibrium (purple arrows between no. 6 and 7), in which Glc7 appears to counteract Snf1 phosphorylation of Reg1 and promote its own dissociation from active kinase complexes, is at the heart of this regulatory duel. Sip5 may stabilize the Reg1/Snf1 interaction in no. 6 (not shown). Hxk2 interacts with Reg1 (not shown), and Hxk2 may respond to PKA signaling by dimerizing and mediating the switch in Glc7 phosphatase substrate selection (from Reg1 [no. 7] to Snf1 [no. 1]; see the text).