FIG. 1.
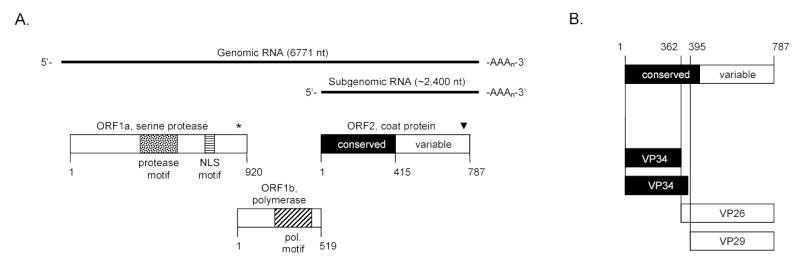
Genomic organization and morphogenesis of HAstV-1 prototype (Oxford strain). (A) Genomic and subgenomic RNA are indicated by the solid black line with their length indicated in nucleotides (nt). The open reading frames (ORF) along with their protein products are shown as rectangles. ORF1a encodes a 3C-like serine protease motif (stippled box) along with a nuclear localization (NLS) motif (horizontally striped box). The asterisk (*) denotes the ribosomal frameshifting signal. ORF1b encodes the viral polymerase and the polymerase motif is indicated (box with diagonal lines). ORF2 encodes the coat protein. The first half of the coat protein sequence (amino acids 1 to 415, heavily shaded) is highly conserved among all known members of the Astroviridae whereas the last half of the coat protein (amino acids 416–787, no shading) is highly variable. The arrowhead (▾) indicates the approximate location of putative caspase cleavage site(s) conserved among astrovirus capsid proteins (42). The numbers below the rectangles refer to length of the protein products in amino acids. (B) Cleavage cascade of the coat protein precursor as described in the text. Trypsin treatment cleaves the particle into its mature products of VP34 (of which two species are predicted), VP26 and VP29.
