Fig. 4.
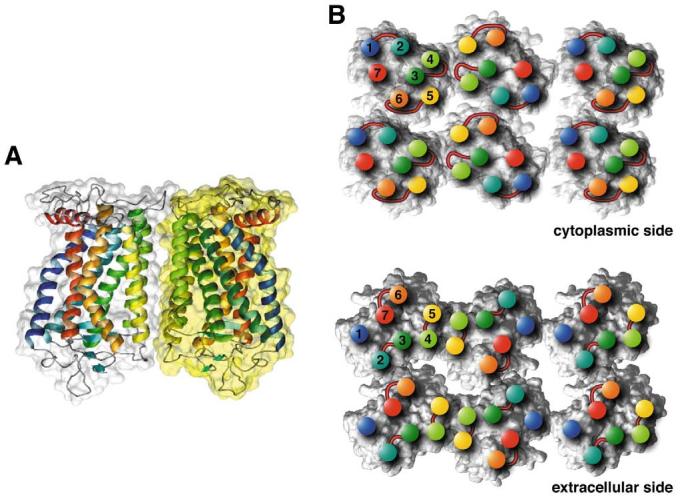
Model for the packing arrangement of rhodopsin molecules within the paracrystalline arrays of native disc membranes (based on [45,46]). A: Side view of a rhodopsin dimer, the building block of the paracrystal. Contacts between monomers are formed by the transmembrane helices IV (yellow-green) and V (yellow). Most of the interacting residues are located on the cytoplasmic loop between helices H-III and H-IV, and on the C-terminal region. Other interaction sites are located within the membrane. Both areas contain hydrogen bonds as well as interactions of hydrophobic nature. B: Cytoplasmic side (top) and extracellular side (bottom) of rhodopsin oligomers. Positions of helix ends are marked by colored discs and the corresponding helix numbers. Extracellular and cytoplasmic loops are drawn schematically at the corresponding locations. Contacts between dimers are created entirely by the intracellular loop between H-V and H-VI from one monomer with the loop between H-I and H-II and the C-terminal residues from the partner monomer. Only half of a second row of rhodopsins is shown. The contact between double rows is created mainly by hydrophobic residues from H-I close to the extracellular side. The lipid molecules initially included in the model for molecular dynamics are not displayed.
