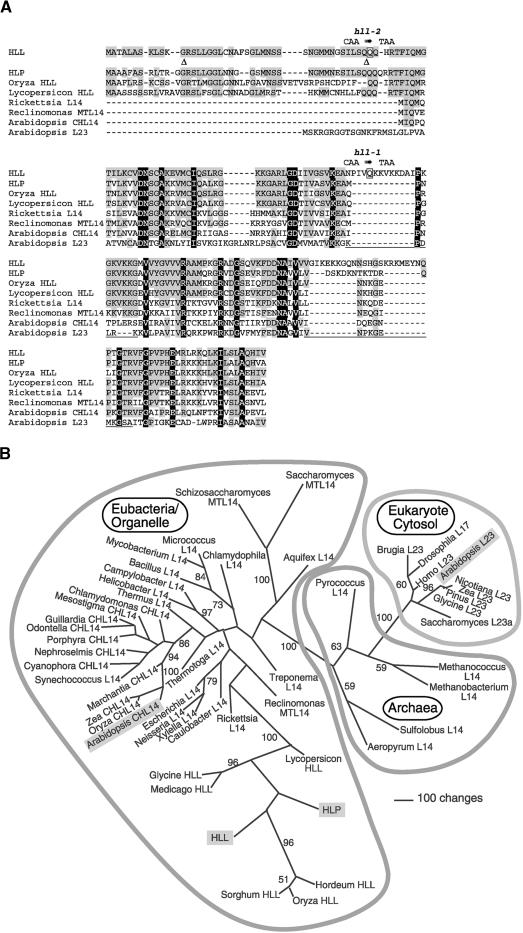
Figure 3.
Sequence Alignment of Proteins Similar to HLL and Phylogenetic Analysis of L14 Homologs from Diverse Lineages.
(A) Deduced protein sequences of HLL and HLP, aligned with those encoded by similar genes from other plants (rice [Oryza sativa] and tomato [Lycopersicon esculentum]), a eubacterial ancestor of mitochondria (Rickettsia prowazekii), the Reclinomonas americana mitochondrion (Reclinomonas MTL14), the Arabidopsis chloroplast (Arabidopsis CHL14), and the Arabidopsis cytosolic L14 homolog (Arabidopsis L23). Amino acids with black backgrounds are identical in all illustrated proteins, and gray shading denotes identity to HLL. The positions of introns in the HLL gene are indicated by triangles. Amino acids altered in the hll-1 and hll-2 alleles are shown in boxes with the DNA sequence change above. Underlines correspond to the locations of loops in the protein structure of the corresponding regions of Bacillus stearothermophilus L14 (Ramakrishnan et al., 1995).
(B) One of eight most parsimonious trees resulting from the analysis of HLL and related proteins. A selection of L14 and L23 proteins from a variety of organisms (and subcellular compartments: CH, chloroplast; MT, mitochondria; all others, cytosolic/cytoplasmic) were analyzed. Insertions and deletions that were not phylogenetically informative were omitted from analysis. The illustration shows an unrooted phylogram of one of the eight shortest trees (13,291 steps) that resulted from 500 random taxon addition replications of a heuristic search on random starting trees using the tree bisection reconnection algorithm for branch swapping. The other trees of equivalent length differed from this tree only in relationships among plant L23 proteins and among the taxa of the three-member clade that includes the Synechococcus sequence. Values from 500 bootstrap replicates (same search settings as described above, except that five random addition trials were performed for each replicate) are shown for those branches that obtained >50% support. Outlines indicate phylogenetic domains of the sequence sources. Sequences from Arabidopsis are highlighted with shaded boxes. For accession numbers see Methods.