Abstract
Burkholderia mallei lipopolysaccharide (LPS) has been previously shown to cross-react with polyclonal antibodies raised against B. pseudomallei LPS; however, we observed that B. mallei LPS does not react with a monoclonal antibody (Pp-PS-W) specific for B. pseudomallei O polysaccharide (O-PS). In this study, we identified the O-PS biosynthetic gene cluster from B. mallei ATCC 23344 and subsequently characterized the molecular structure of the O-PS produced by this organism.
Burkholderia mallei is a gram-negative bacterium responsible for a disease known as glanders in solipeds and occasionally in humans (3, 8, 13). The factors involved in the pathogenesis of B. mallei infection remain relatively poorly defined at the molecular level. A previous study that identified a polysaccharide gene cluster in B. mallei showed that B. mallei lipopolysaccharide (LPS) cross-reacts with polyclonal antibodies raised against the LPS of Burkholderia pseudomallei, a closely related organism responsible for a disease known as melioidosis (6). In the present study, we investigated the LPS profiles of B. mallei strains, identified the gene cluster responsible for O polysaccharide (O-PS) biosynthesis in B. mallei ATCC 23344, and determined the physical structure of the B. mallei ATCC 23344 O-PS. Additionally, we showed that the O-PS moiety of B. mallei LPS is required for resistance to the bactericidal action of serum. Finally, we identified the presence of insertion sequences in two strains of B. mallei that disrupt the expression of O-PS.
Analysis of LPS profiles of B. mallei strains.
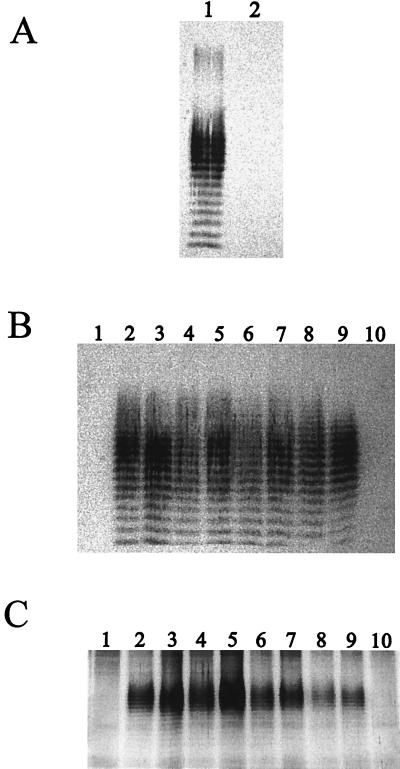
The strains and plasmids used in this study are shown in Table 1. The first goal of this study was to assess the LPS profiles of B. mallei strains. Initially, we performed Western blot analysis of B. mallei ATCC 23344 whole-cell lysates with polyclonal rabbit sera raised against a B. pseudomallei bovine serum albumin (BSA)-O-PS conjugate as well as with a B. pseudomallei O-PS-specific MAb (Pp-PS-W) according to a previously described protocol (1, 2). As shown in Fig. 1A, B. mallei ATCC 23344 reacted with the anti-LPS polyclonal sera, resulting in a typical LPS banding pattern; however, the B. pseudomallei O-PS-specific MAb (Pp-PS-W) did not react. This indicated that differences exist between B. mallei and B. pseudomallei O-PS. We further assessed the LPS profiles of 10 different B. mallei strains (Fig. 1B). By using Western blot analysis, we showed that 8 of the 10 strains assessed bound the anti-LPS polyclonal sera and displayed typical LPS banding patterns. In contrast, however, two strains, NCTC 120 and ATCC 15310, did not bind the anti-LPS polyclonal sera, as indicated by the absence of bands (Fig. 1B). In order to confirm that the O-PS moiety was absent rather than a different type of O-PS, silver stain analysis was employed. Figure 1C shows the silver stain results confirming that both of these strains lacked O-PS moieties.
TABLE 1.
Bacterial strains and cosmids or plasmids used in this study
| Strain, cosmid, or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| SM10 | Mobilizing strain: transfer genes of RP4 integrated into the chromosome; Kmr Sms | 11 |
| TOP10 | High-efficiency transformation strain with blue/white screening; Aps Kms | Invitrogen |
| HB101 | Serum-sensitive strain | 7 |
| B. mallei | ||
| NCTC 120 | USAMRIIDa | |
| NCTC 10248 | Human isolate | USAMRIID |
| NCTC 10229 | USAMRIID | |
| NCTC 10260 | Human isolate | USAMRIID |
| NCTC 10247 | USAMRIID | |
| ATCC 23344 | Human isolate; Pmr Gms | USAMRIID |
| NCTC 3708 | Mule isolate | USAMRIID |
| NCTC 3709 | Horse isolate | USAMRIID |
| ATCC 10399 | USAMRIID | |
| ATCC 15310 | USAMRIID | |
| PB100 | ATCC 23344::pGSV3008; Pmr Gmr | This study |
| B. pseudomallei 1026b | Clinical isolate; Gmr Kmr Smr Pmr Tps | 4 |
| Cosmids | ||
| pScosBC1 | Broad-host-range cosmid cloning vector based on pSuperCos1; Apr Tpr | 12 |
| p1C3 | pScosBC1 from ATCC 23344 library with a 23-kb fragment containing the O-PS biosynthetic gene cluster | This study |
| p2B5 | pScosBC1 from ATCC 23344 library with a 27-kb fragment containing the O-PS biosynthetic gene cluster | This study |
| Plasmids | ||
| pUC19 | Cloning vector with blue/white selection; Apr | 14 |
| pGSV3008 | pGSV containing a 379-bp EcoRI fragment from pDD3008, contains internal fragment from the wcbB gene; Gmr | 6 |
U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, Md.
FIG. 1.
(A) Western blot analysis of B. mallei ATCC 23344. Proteinase K-treated whole-cell lysates were used. In lane 1, the primary antibody used was a 1/2,000 dilution of polyclonal antisera raised against a B. pseudomallei BSA-O-PS conjugate, and in lane 2, the primary antibody used was a 1/2,000 dilution of the B. pseudomallei O-PS-specific MAb (Pp-PS-W). (B) Western blot profiles of proteinase K-treated whole-cell lysates of B. mallei strains. The primary antibody used was polyclonal sera raised against a B. pseudomallei BSA-O-PS conjugate. Lanes: 1, NCTC 120; 2, NCTC 10248; 3, NCTC 10229; 4, NCTC 10260; 5, NCTC 10247; 6, ATCC 23344; 7, NCTC 3708; 8, NCTC 3709; 9, ATCC 10399; and 10, ATCC 15310. (C) Silver stain analysis of proteinase K-treated whole-cell lysates of B. mallei strains. Lanes: 1, NCTC 120; 2, NCTC 10248; 3, NCTC 10229; 4, NCTC 10260; 5, NCTC 10247; 6, ATCC 23344; 7, NCTC 3708; 8, NCTC 3709; 9, ATCC 10399; and 10, ATCC 15310.
Identification and characterization of B. mallei ATCC 23344 O-PS biosynthetic gene cluster.
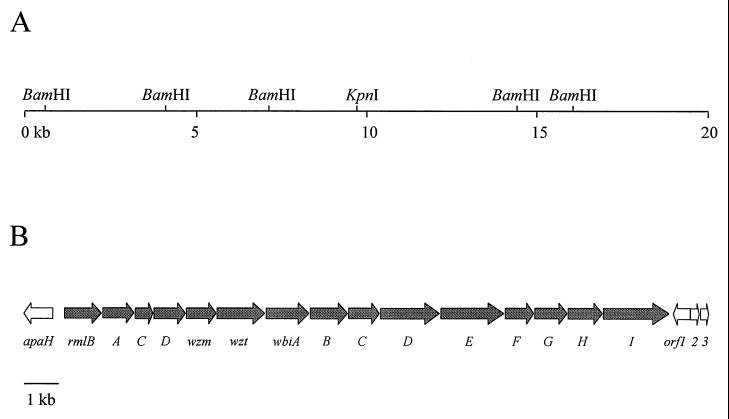
In order to investigate the genes responsible for O-PS biosynthesis in B. mallei, we constructed a cosmid library by using B. mallei ATCC 23344 genomic DNA and the cosmid pScosBC1 by using a previously described protocol (12). Colony hybridizations were then performed with a 1.1-kb DNA fragment containing the recently identified B. mallei wbiA gene (P. Brett, M. Burtnick, and D. Woods, unpublished data). Six positive cosmid clones were obtained. Based on the BamHI-KpnI restriction patterns obtained, two cosmid clones, 1C3 and 2B5, were predicted to harbor the entire B. mallei O-PS gene cluster. Sequence analysis resulted in 19,918 bp of contiguous sequence containing the entire B. mallei O-PS biosynthetic gene cluster with an IS407-like insertion sequence element at the 3′ end.
The first 18,738 bp of the B. mallei DNA sequence contained 16 predicted ORFs that were identical to those previously defined as the O-PS biosynthetic gene cluster in B. pseudomallei (Fig. 2) (5). Sequence alignment of the B. pseudomallei and B. mallei O-PS biosynthetic regions revealed 99% identity at the nucleotide level. The genes comprising the B. mallei O-PS biosynthetic operon were named as per the identical genes found in B. pseudomallei (5).
FIG. 2.
Restriction and genetic maps of the B. mallei O-PS biosynthetic gene cluster. (A) Restriction map. The horizontal line represents the B. mallei DNA insert of cosmid 2B5. The locations of BamHI and KpnI cleavage sites used for subcloning are shown. Two additional BamHI sites at the 5′ and 3′ ends of 2B5, which were part of the pScosBC1 vector, are not shown. (B) Genetic map. The location and direction of transcription of the genes are represented by arrows, and the gene names are shown below. The gray arrows indicate the genes involved in O-PS biosynthesis based on homology to the B. pseudomallei O-PS biosynthetic operon.
Physical characterization of B. mallei O-PS moieties.
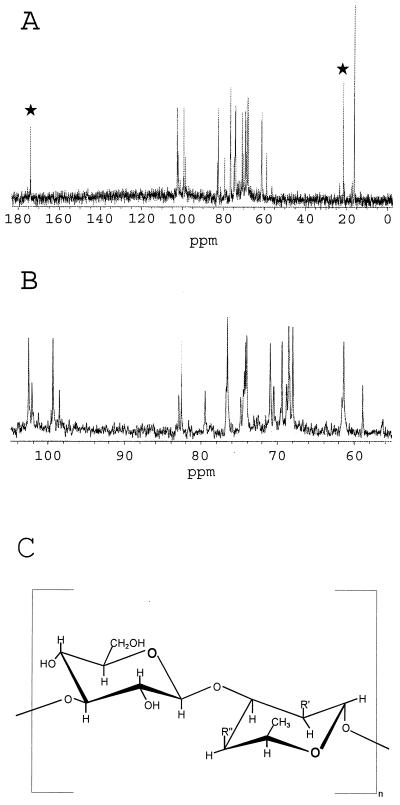
In order to structurally analyze the B. mallei O-PS structure, it was necessary to construct a B. mallei strain unable to produce capsular polysaccharide (CPS), because CPS copurifies with LPS. The suicide vector pGSV3008 was employed as previously described to construct B. mallei PB100, a derivative of ATCC 23344 that does not produce CPS (6). The O-PS was purified as previously described for B. pseudomallei. Figure 3 shows 13C nuclear magnetic resonance (13C-NMR) analysis (Complex Carbohydrate Research Center, University of Georgia, Athens) results demonstrating that the B. mallei O-PS backbone is similar to that previously described for B. pseudomallei O-PS, a heteropolymer of repeating d-glucose and l-talose (9, 10). However, changes are apparent in the O-acetylation pattern of the B. mallei l-talose residue in comparison to that of B. pseudomallei. Similar to B. pseudomallei O-PS, B. mallei O-PS demonstrates the presence of O-acetyl or O-methyl substitutions at the 2′ position of the talose residue. In contrast, B. mallei O-PS is devoid of an O-acetyl group at the 4′ position of the talose residue. Thus, the structure of B. mallei O-PS is best described as 3)-β-d-glucopyranose-(1,3)-6-deoxy-α-l-talopyranose-(1-, in which the talose residue contains 2-O-methyl or 2-O-acetyl substituents. Recent studies indicate that the presence of 4-O-acetyl groups on the talose residues of B. pseudomallei O-PS is due to an O-acetylation locus unlinked to the previously described O-PS biosynthetic operon (Brett et al., unpublished). If this is the case, then the unlinked locus responsible for O-acetylation is either not present or is nonfunctional in B. mallei strains. The presence or absence of O-acetyl groups on the O-PS moieties may have consequences when O-PS is considered as a component of a vaccine that protects against both B. mallei and B. pseudomallei.
FIG. 3.
13C-NMR analysis of B. mallei PB100 O-PS. (A) O-Acetyl peaks are indicated by stars. (B) Expanded view of the region running from 100 to 60 ppm. (C) Structure of B. pseudomallei and B. mallei O-PS. In B. pseudomallei, in 33% of the talose residues, R′ = O-methyl and R" = O-acetyl, and in 66% of the talose residues, R′ = O-acetyl and R" = OH. In B. mallei, R′ = O-acetyl or O-methyl and R" = OH.
B. mallei survives in 30% NHS, and serum-sensitive strains lack the O-PS moiety of LPS.
The ability of B. mallei ATCC 23344 to grow in the presence of 30% normal human serum (NHS) was initially assessed with a serum bactericidal assay (5) in which viable counts were determined at 2, 4, 8, and 18 h. B. mallei ATCC 23344 was shown to survive in the presence of 30% NHS over the course of 18 h (Fig. 4A). Serum-resistant B. pseudomallei 1026b and serum-sensitive Escherichia coli HB101 were employed as controls.
FIG. 4.
Serum bactericidal assays with B. mallei strains. (A) Thirty percent NHS killing assay in which viable counts were determined at the 2-, 4-, 8-, and 18-h time points. B. pseudomallei 1026b (▴), B. mallei ATCC 23344 (⧫), and E. coli HB101 (•). (B) Thirty percent NHS killing assay in which viable counts were determined following a 2-h incubation at 37°C. Control tubes containing phosphate-buffered saline (PBS) are shown as white bars, and experimental tubes containing 30% NHS are shown as gray bars. Error bars indicate standard deviations.
In order to assess the role of B. mallei O-PS in serum resistance, NHS bactericidal assays (5) were performed with B. mallei ATCC 23344 and B. mallei NCTC 120 and ATCC 15310, the two strains lacking O-PS. B. mallei ATCC 23344 remained resistant to the killing action of 30% NHS, while NCTC 120 and ATCC 15310 were completely killed following a 2-h incubation in 30% NHS (Fig. 4B). The other seven B. mallei strains used in this study possessed intact O-PS moieties and were resistant to the bactericidal action of 30% NHS (data not shown). These results suggested that B. mallei O-PS moieties play a crucial role in the serum resistance of this organism: this correlates well with previous studies demonstrating that B. pseudomallei O-PS is required for serum resistance (5).
Identification of insertion sequence IS407 in the O-PS biosynthetic gene clusters of B. mallei NCTC 120 and ATCC 15310.
In order to determine if the O-PS biosynthetic gene clusters of NCTC 120 and ATCC 15310 had been disrupted, we chose to individually PCR amplify each gene present in this cluster. Deoxyoligonucleotide primers were designed outside of the 5′ and 3′ ends of each gene. B. mallei ATCC 23344 chromosomal DNA was used a control as an indicator of the size of a wild-type copy of each gene. Alterations were observed in the wbiE PCR product from NCTC 120 and in the wbiG PCR product from ATCC 15310. The PCR products obtained in both cases were approximately 1.5 kb larger than those obtained with ATCC 23344 genomic DNA (data not shown). Cloning and sequence analysis of the NCTC 120 wbiE and ATCC 15310 wbiG PCR products revealed the presence of insertion sequences within these two genes. In NCTC 120, an IS407-like element was located after nucleotide 13615 of the O-PS operon in the wbiE gene. In ATCC 15310, an IS407-like element was located following nucleotide 15107 of the O-PS operon in the wbiG gene. It is likely that the presence of insertion elements in the O-PS biosynthetic gene clusters of B. mallei NCTC 120 and ATCC 15310 is responsible for the loss of expression of O-PS in these two strains. DeShazer et al. have previously demonstrated the presence of an IS407-like element (termed “IS407A”) at the 3′ end of the CPS gene cluster and have shown that this element is active in B. mallei (6). The data presented in this paper certainly support the view that IS407 is functionally active in B. mallei.
Acknowledgments
This work was funded by the Department of Defense contract no. DAMD 17-98-C-8003 and CIHR MOP 31343. M.N.B. is the recipient of an Alberta Heritage Foundation for Medical Research Studentship Award.
We are grateful to Patricia Baker and Francois Becotte for excellent technical assistance. We thank David DeShazer for providing us with the plasmid pGSV3008.
REFERENCES
- 1.Brett, P. J., and D. E. Woods. 1996. Structural and immunological characterization of Burkholderia pseudomallei O-polysaccharide-flagellin protein conjugates. Infect. Immun. 64:2824–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryan, L., S. Wong, D. Woods, D. Dance, and W. Chaoagul. 1994. Passive protection of diabetic rats with antisera specific for the polysaccharide portion of lipopolysaccharide from Pseudomonas pseudomallei. Can. J. Infect. Dis. 5:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2000. Laboratory acquired human glanders—Maryland May 2000. Morb. Mortal. Wkly. Rep. 49:532–533. [PubMed] [Google Scholar]
- 4.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeShazer, D., P. J. Brett, and D. E. Woods. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30:1081–1100. [DOI] [PubMed] [Google Scholar]
- 6.DeShazer, D., D. M. Waag, D. L. Fritz, and D. E. Woods. 2001. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb. Pathog. 30:253–269. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez, R. C., and A. A. Weiss. 1994. Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect. Immun. 62:4727–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howe, C., and W. Miller. 1948. Human glanders: report of six cases. Ann. Intern. Med. 26:93–115. [DOI] [PubMed] [Google Scholar]
- 9.Knirel, Y. A., N. A. Paramonov, A. S. Shashkov, N. K. Kochetkov, R. G. Yarullin, S. M. Farber, and V. I. Efremenko. 1992. Structure of the polysaccharide chains of Pseudomonas pseudomallei lipopolysaccharides. Carbohydr. Res. 233:185–193. [DOI] [PubMed] [Google Scholar]
- 10.Perry, M. B., L. L. MacLean, T. Schollaardt, L. E. Bryan, and M. Ho. 1995. Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect. Immun. 63:3348–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784–791. [Google Scholar]
- 12.Sokol, P. A., P. Darling, D. E. Woods, E. Mahenthiralingam, and C. Kooi. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5- oxygenase. Infect. Immun. 67:4443–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson, L. 1981. Glanders: medicine and veterinary medicine in common pursuit of a contagious disease. Med. Hist. 25:363–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. [DOI] [PubMed] [Google Scholar]