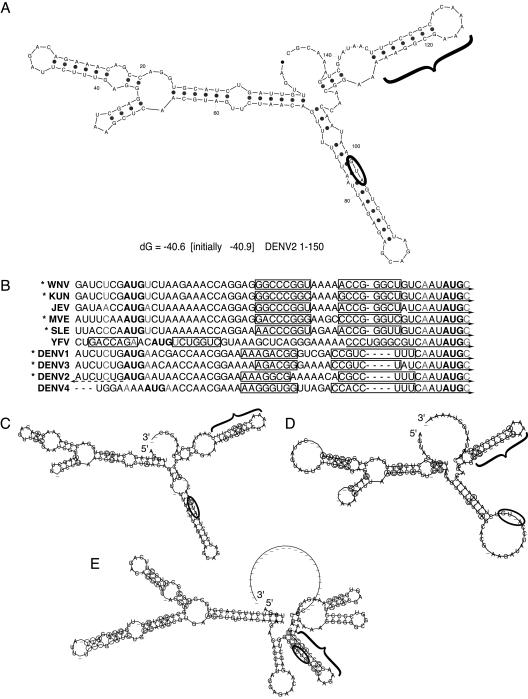
FIG. 2.
A conserved hairpin element is predicted among mosquito- and tick-borne flaviviruses. (A) The RNA secondary structure of the first 150 nt of DENV2 was predicted by mfold; the start codon is circled, and the cHP is indicated by a bracket. (B) Alignment of start codons and the first in-frame AUG of mosquito-borne flaviruses, with predicted cHP stem regions outlined by rectangles. The −3 and +4 positions are indicated in gray letters. Viruses with a poor initiation context are indicated by asterisks. 5′ CS regions are indicated by solid arrows. The DENV2 5′ UAR region is indicated by a dashed arrow. (C) Phylogenetic consensus structure based on aligned sequences of DENV1, DENV2, DENV3, and DENV4 as computed by RNAalifold, with the start codon circled and the cHP indicated with a bracket. Covariant residues are circled. (D) Phylogenetic consensus structure of the mosquito-borne Japanese encephalitis serogroup viruses, WNV, Kunjin virus, JEV, St. Louis encephalitis virus, and Murray valley encephalitis virus, as in panel C. (E) Phylogenetic consensus structure of the tick-borne viruses: tick-borne encephalitis, Omsk hemorrhagic fever, Kyasanur Forest disease, and Powassan viruses, as in panel C.