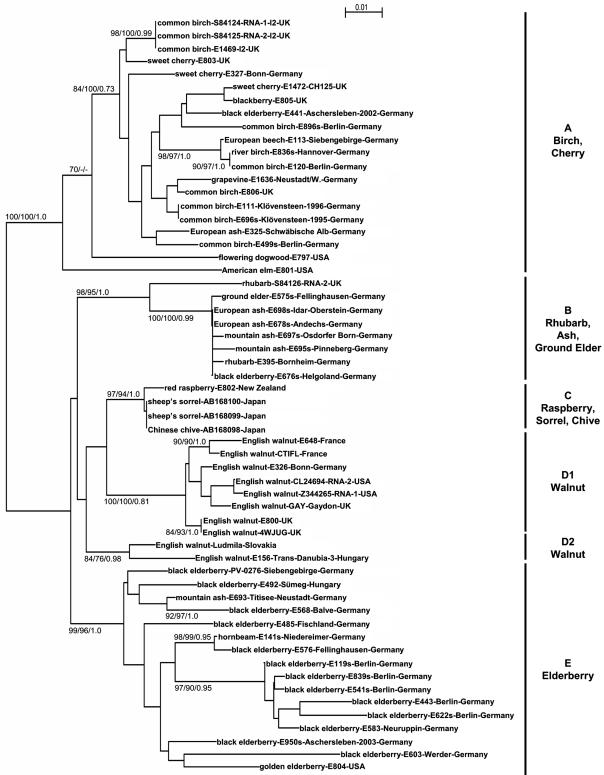
FIG. 1.
Phylogenetic tree reconstructed from the nucleotide sequence of a 3′-terminal genomic fragment (375 bp) of the cherry leaf roll virus genome amplified from original host plants or from isolates recovered from various hosts. Details about isolates are shown in Table 1. All data obtained from the EMBL nucleotide database are indicated by accession numbers. Data analysis and tree construction were done by neighbor-joining, maximum likelihood, and Bayesian analyses using the CLUSTALX, Phylip, and MrBayes programs. Bootstrap values (n = 1,000) or probability estimate values larger than 70% are indicated at branch nodes for neighbor-joining/maximum-likelihood/Bayesian analysis. Major phylogenetic groups are indicated by a bold line on the right.