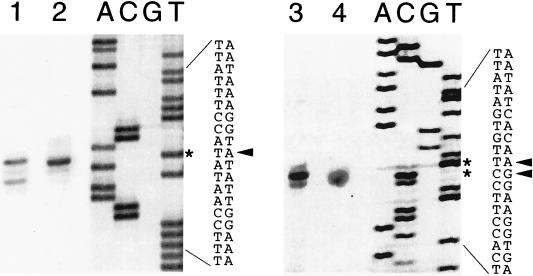
FIG. 1.
Mapping of in vivo and in vitro transcription start sites for sinIR promoters. Start sites were determined by reverse transcriptase extension of 32P-end-labeled primers by using in vivo RNA isolated from IS75 (16), grown in NSM, and harvested at T0 and in vitro RNA isolated from transcription reaction mixtures. B. subtilis RNA polymerase holoenzyme containing ςA was allowed to transcribe pSM109 (16), a plasmid containing sinIR P1 and P3, with nonradioactive ribonucleoside triphosphates, and RNA was isolated from the reaction mixtures. After radioactive primers were annealed to the RNAs and after extension with reverse transcriptase, the samples were separated by electrophoresis on 6% polyacrylamide gels, followed by autoradiography. Dideoxy sequencing gel lanes 1 and 2 show the primer extension products obtained from in vitro RNA (lane 1) and in vivo RNA (lane 2) using primer OSIN-1. The dideoxy sequencing ladder adjacent to lane 2 was obtained with the same primer and pSM179. The nucleotide sequence with the arrow pointing to the derived transcriptional start sites, also indicated by asterisks, is presented in the adjacent lanes. Lanes 3 (in vitro RNA) and 4 (in vivo RNA) show the equivalent primer extension products with primer P3-1, which corresponds to a region ca. 60 nt downstream from P3. Adjacent to lane 4 is the sequencing ladder obtained from pSM109 (16) with primer P3-1. The DNA sequence and derived transcription start sites are also presented.