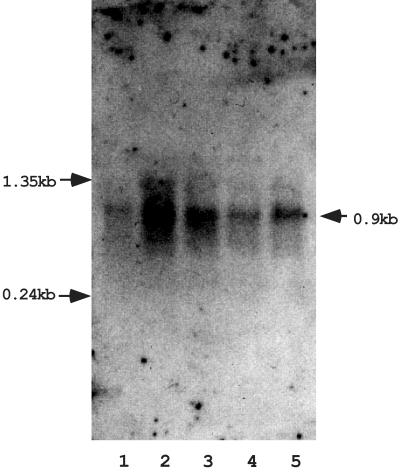
FIG. 1.
Northern blot analysis of the conjugation-regulating gene 71ORF2. Total RNAs were prepared from mating mixtures at various times during conjugation. Clone #71 DNA (6 kbp) was used as a probe. Matings carried out for the purpose of preparation of RNA were conducted as follows. After overnight growth, 5 ml of the donor strain was added to 25 ml of fresh THB in a 50-ml centrifugation tube and incubated on a roller at 37°C for 4 h. Concurrently, 5 ml of an overnight culture of the recipient strain was mixed with 25 ml of fresh THB in a 100-ml flask and incubated with gentle shaking at 37°C for 4 h. Then 2.5 ml of the recipient culture was added to the donor culture to initiate the mating. Mating was conducted at 37°C for the desired time and was then terminated by addition of chloramphenicol at a final concentration of 200 μg/ml. The mating mixture was then placed on ice for 15 min and harvested by centrifugation. The pellet was suspended in 600 μl of lysis buffer (50 mM glucose, 25 mM Tris-HCl; pH 8.0). Fifty microliters of a lysozyme solution (100 mg/ml) was added, and the cells were incubated at 37°C for 30 min. To lyse the cells, 800 μl of TRIzol (Invitrogen, Inc., Carlsbad, Calif.) and 80 μl of chloroform were added, and the mixture was vortexed for 20 s. The mixture was kept on ice for 5 min and spun for 15 min at 4°C. The aqueous phase was transferred to a new tube and extracted twice with phenol-chloroform-isoamyl alcohol. Nucleic acid was recovered by ethanol precipitation, and the pellet was suspended in 100 μl of DNase I buffer. DNA was digested by adding 30 U of RNase-free DNase I (10 U/μl; Roche Diagnostics GmbH, Mannheim, Germany) and incubating the preparation at 37°C for 2 h. The RNA was treated with phenol-chloroform, recovered by ethanol precipitation, and suspended in 50 μl of diethyl pyrocarbonate-treated distilled water. Northern blotting was performed by using standard protocols (2, 22, 24). RNA samples were electrophoresed on a 1% agarose gel with morpholinepropanesulfonic acid (MOPS)-formaldehyde buffer and were transferred to a nylon membrane (Roche Diagnostics GmbH) by the conventional capillary method. Hybridization and signal detection were performed by using a DIG DNA labeling and detection kit (Roche Diagnostics GmbH) according to the manufacturer's protocols. EasyHyb (Roche Diagnostics GmbH) was used as the prehybridization and hybridization solutions, and CSPD (Roche Diagnostics GmbH) was used as a substrate for chemical luminescence. Lane 1, OG1X(pMG1) donor cells before conjugation; lane 2, 20 min; lane 3, 40 min; lane 4, 80 min; lane 5, 160 min. The arrows on the left indicate the positions of the RNA molecular weight markers (Invitrogen, Inc.). The arrow on the right indicates the estimated position of the band.