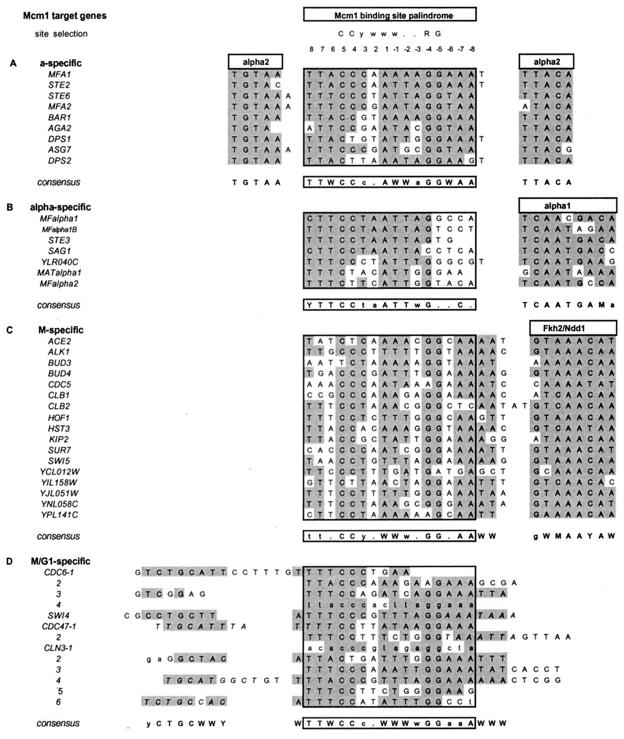
FIG. 1.
Binding sites of Mcm1-containing complexes. Compilation of Mcm1 binding sites in the promoters of four different classes of Mcm1 target genes based on previous studies (32, 44, 52, 54). The minimal Mcm1 binding site identified by site selection is shown at the top. Below are the in vivo binding sites for four different classes of Mcm1 target genes, and the 16-bp Mcm1 binding site is boxed. Positions fitting the consensus sequence for each group are shaded, and the consensus is shown below. Bases written in uppercase depict those which are more than 75% conserved; bases in lowercase show positions identical in at least half the target genes. W = A or T, K = G or T, M = A or C, Y = C or T, R = A or G, and a dot indicates any base. Boxes above each flanking homology indicate the proteins which are known to bind to those sites and confer regulatory specificity to the complex. The M/G1-specific genes are shown in section D, and all the sequences that are protected from DNase I cleavage (see Fig. 2 and 3) are shown in capital letters. Other potential ECB elements and a few other residues that are not protected from DNase I are shown in lowercase for purposes of comparison. Italic letters indicate that footprints were obtained only on the opposite strand to that shown. The sequence flanking the Mcm1 binding site in the M/G1-specific promoters is aligned to show a region of limited sequence homology that is protected from DNase I cleavage.