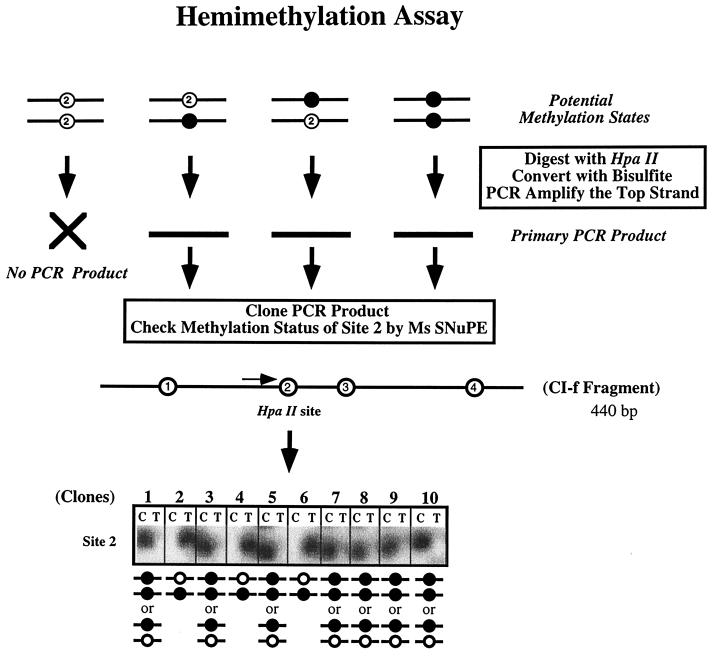
FIG. 4.
Detection and quantitation of hemimethylation in individual cloned DNA molecules. The experimental approach used to detect hemimethylation in the ES cells consists of precutting genomic DNA with HpaII, followed by bisulfite treatment, then cloning individual PCR products, and assessing the methylation status of site 2 by Ms-SNuPE analysis. Shown are typical Ms-SNuPE results in which a signal in the T lane for a clone indicates hemimethylation, with the top strand being unmethylated. The presence of a signal in the C lane indicates that the site may be either hemimethylated (i.e., the bottom strand is unmethylated) or fully methylated. The distribution between these two scenarios is determined by calculation, assuming that there is an equal probability of hemimethylation of either the top or the bottom strand.