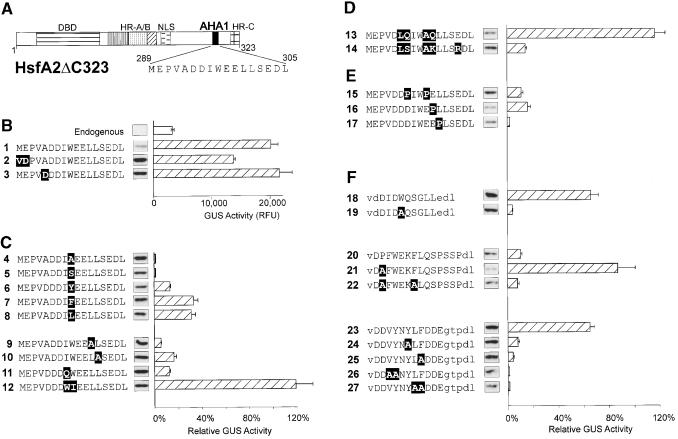
Figure 2.
Mutational Analysis of AHA Motifs in the Context of HsfA2ΔC323.
The activator potential of the indicated HsfA2ΔC323 mutants was tested in tobacco protoplasts cotransformed with the phsp17GUS reporter plasmid. The basic structure (A) and the partial amino acid sequences of the AHA1 motif in groups (B) to (F) identify the individual constructs. Point mutations are indicated with white letters on black background. For group (F) with heterologous AHA motifs replacing the AHA1 motif of HsfA2, the capital letters represent the amino acid sequences of the indicated parts of HsfA1 (constructs 18 to 22; see details given in Figure 3B) and yeast Gal4p (amino acid residues 862 to 872), respectively. Lowercase letters in the sequence data indicate flanking sequences, including the VD (SalI site) and DL (BglII site) motifs used for linker insertion. The relative GUS activities are indicated by the hatched bars. They refer to the activity of the basal constructs (B) set as 100%. Results with constructs 4, 5, and 7 to 10 refer to the activity of construct 1, results with constructs 18 to 27 refer to construct 2, whereas constructs 6 and 11 to 17 refer to construct 3. The expression levels are indicated by the signals from protein gel blot analysis shown between the AHA sequence and the corresponding GUS activity bar. Error bars indicate the absolute deviation from the mean values. DBD, DNA binding domain; NLS, nuclear localization signal; RFU, relative fluorescence units.