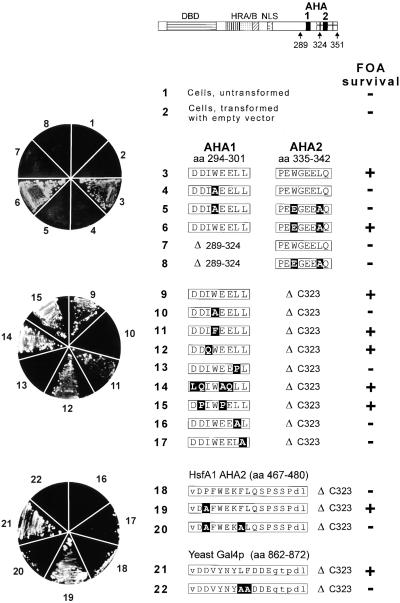
Figure 4.
HsfA2 Forms with an Active CTAD Support Growth of Yeast Strains in Which the Endogenous HSF1 Gene Is Disrupted.
Cells of the yeast strain RSY4 containing a chromosomal disruption of the HSF1 gene (Boscheinen et al., 1997) and a copy of the yeast HSF1 gene on a URA3 plasmid were transformed with a TRP1 plasmid coding for the indicated form of HsfA2. The URA3 plasmid was eliminated by plating on media containing FOA. FOA survival is shown by the pictures of agar plates at left. Details of the HsfA2 cassettes in the yeast vector pMBI are given by the block diagram on top and the sequence information for constructs 3 to 22 (for further explanation, see Figures 1 and 2). Point mutations are printed in white letters on a black background. aa, amino acid residues; DBD, DNA binding domain; HRA/B, hydrophobic heptad repeat region for oligomerization; NLS, nuclear localization signal; (+), survival on FOA plates; (−), no survival.