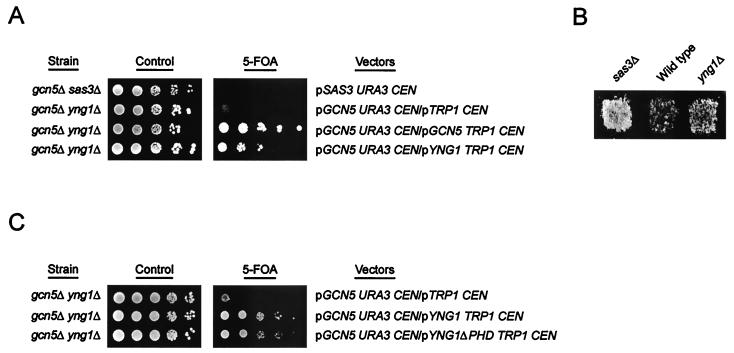
FIG. 4.
Loss of YNG1 disrupts NuA3 function. (A) gcn5Δ yng1Δ strains are synthetically sick. Yeast strain YJW542 (MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 gcn5::HIS3 yng1::HISMX6 pJW218) was transformed with a CEN/ARS TRP1 plasmid alone (pRS314) or with CEN/ARS TRP plasmids encoding Gcn5p or Yng1p (pJW215 and pJW221, respectively), and the resulting transformants were plated on either synthetic complete medium (control) or synthetic complete medium with 5-FOA. Strain YJW134 (MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 gcn5::HIS3 sas3::HISMX6 pJW214) was used as a control. (B) Deletion of YNG1 restores silencing to strains with mutations in HMR-E. Wild-type (JRY2069; MATα HMRa-e∗∗ ade2-101 his3 lys2 tyr1 ura3-52) and yng1Δ (YJW548; MATα HMRa-e∗∗ ade2-101 his3 lys2 tyr1 ura3-52 yng1::HISMX6) strains were assayed for mating efficiency, with JRY2726 (MATa his4) as a tester strain. Strain YJW547 (MATα HMRa-e∗∗ ade2-101 his3 lys2 tyr1 ura3-52 sas3::HISMX6) was used as a positive control. (C) Yeast strains YJW542 (MATa his3Δ200 leu2Δ1 ura3-52 trp1Δ63 gcn5::HIS3 yng1::HISMX6 pJW218) was transformed with a CEN/ARS TRP1 plasmid alone (pRS314) or with CEN/ARS TRP1 plasmids encoding Yng1p or Yng1ΔPHD (pJW221 and pJW222, respectively), and the resulting transformants were plated on either synthetic complete medium (control) or synthetic complete medium with 5-FOA.