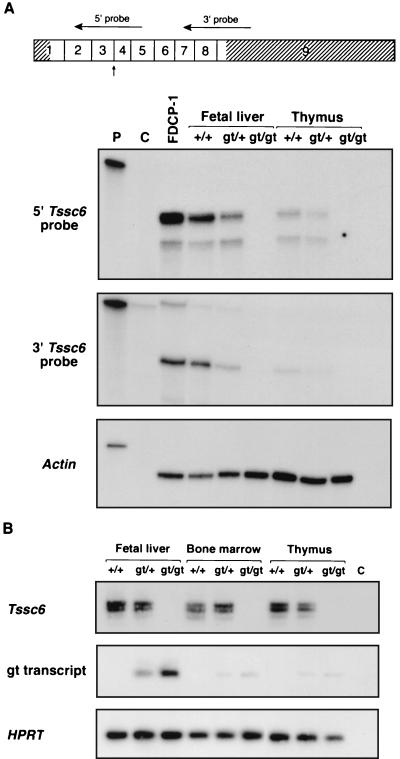
FIG. 2.
The gene trap insertion in Tssc6 results in a null allele. (A) RNase protection analysis of RNA prepared from the fetal liver and thymus of Tssc6gt/gt mice and littermates. At the top of the panel, the Tssc6 cDNA is shown, with the nine exons indicated by boxes. The 5′ and 3′ untranslated regions are hatched. Horizontal arrows indicate the position of the 5′ and 3′ riboprobes. The vertical arrow shows the point of fusion with the gene trap vector. With both 5′ and 3′ riboprobes, protected bands in RNA prepared from the fetal liver and thymus of Tssc6+/+ and Tssc6gt/+ mice are shown, but in RNA from Tssc6gt/gt mice, bands are absent. Lanes: P, undigested, full-length probe; C, probe after RNase digestion; FDCP-1, RNA from FDCP-1 cells, which are known to express Tssc6; +/+, wild-type; gt/+, heterozygous for the Tssc6 gene trap insertion; gt/gt, homozygous for the Tssc6 gene trap insertion. (B) RT-PCR to amplify Tssc6 transcripts and gene trap fusion transcripts from the fetal liver, bone marrow, and thymus of Tssc6gt/gt mice and their littermates. The multiple bands amplified with Tssc6 primers were due to alternate splice variants (26). Hypoxanthine phosphoribosyltransferase transcripts were amplified to estimate the amount of cDNA present in each reaction. Lane C, no cDNA control.