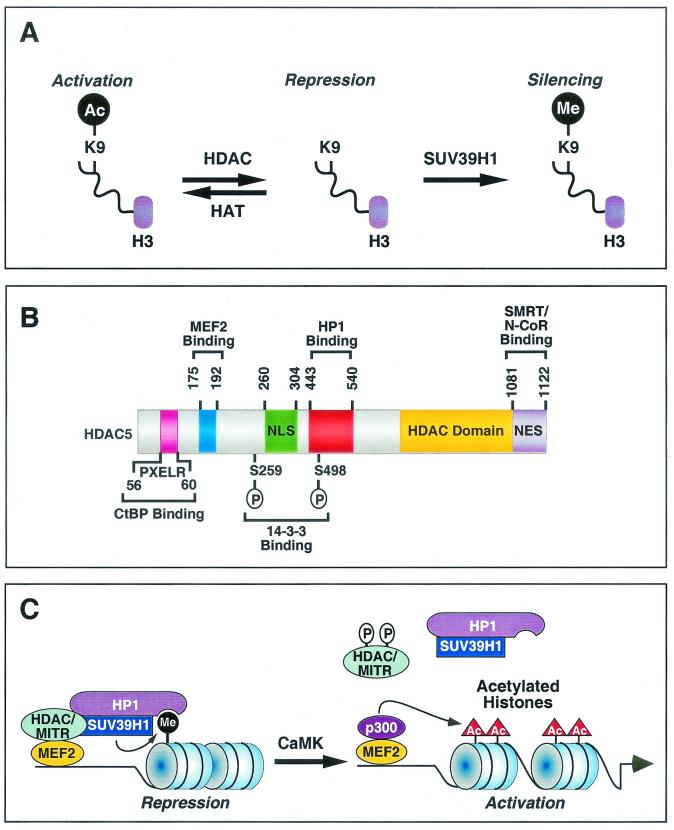
FIG. 6.
Regulation of gene expression by HDAC and MITR-HP1 interactions. (A) Acetylation (Ac) of lysine 9 (K9) on nucleosomal histone H3 by HATs relaxes chromatin structure, resulting in gene activation. In contrast, removal of the acetyl group from K9 by HDACs results in chromatin condensation and gene repression. Deacetylated K9 is a substrate for the HMTase SUV39H1. Methylation of K9 by SUV39H1 further enhances the repressive state of chromatin. (B) Schematic diagram of human HDAC5 showing functional domains and binding regions for cofactors. These functional domains and binding sites are conserved in HDAC4, HDAC7, HDAC9, and MITR, although MITR lacks an HDAC catalytic domain and a nuclear export sequence (NES). NLS, nuclear localization signal.(C) Model for the assembly of a corepressor complex by a class II HDAC or MITR and its disassembly by CaMK signaling. HDAC or MITR, HP1 and the HMTase SUV39H1 form an efficient corepressor complex. Deacetylation of histone tails by HDAC creates substrates for SUV39H1, which is recruited to the target site by the HDAC- or MITR-HP1 complex. Methylation of histone tails by SUV39H1 generates a binding signature for HP1, which further recruits HDACs and SUV39H1 to propagate the repressive effect on the target gene. CaMK phosphorylates class II HDACs and MITR, which results in disruption of MEF2-HDAC or MEF2-MITR interactions and allows p300, which possesses HAT activity, to associate with MEF2 and acetylate regional histones. These events lead to the activation of MEF2 target gene expression. CaMK also dissociates HP1-HDAC and HP1-MITR complexes through an unknown mechanism, providing an efficient means to control histone methylation in response to extracellular cues.