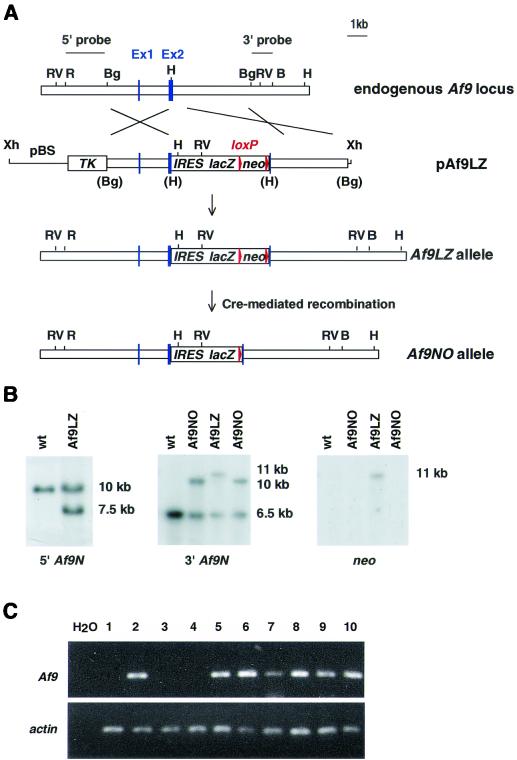
FIG. 1.
Targeting strategy for the generation of Af9 null alleles. (A) A partial genomic map of the mouse Af9 gene around the two exons encoding the first 64 amino acids of mouse Af9 (designated exons 1 and 2) is shown in the top line. Targeting vector pAf9LZ (second line) contained a 6.5-kb genomic BglII fragment, with a 5-kb lacZ-neo cassette inserted into a HindIII site in exon 2. The IRES sequence allowed independent translation of the Escherichia coli lacZ gene, which served as an Af9 gene expression marker. The neo sequence conferred G-418 resistance, allowing positive selection of targeted ES cells. This sequence was flanked by loxP sites to allow subsequent excision by transient expression of Cre recombinase (the starting vector was a gift from K. Douglas and A. Smith). Upstream of the targeting fragment, the herpes simplex virus thymidine kinase gene (TK) cassette allowed negative selection of nonhomologous integration events (62). The regions of homology with the endogenous Af9 gene are indicated. The mutant Af9 allele (Af9LZ) resulting from homologous recombination is illustrated in the third line. The Af9NO allele, resulting from transient expression of Cre recombinase, is illustrated in the fourth line. H, HindIII; B, BamHI; Bg, BglII; R, EcoRI; RV, EcoRV; Xh, XhoI. Blue boxes, exons; red arrowheads, LoxP sites. (B) Filter hybridization analysis of ES clones. A 2-kb nonrepetitive EcoRI-BglII fragment 5′ of the targeted region (A) detected a 10-kb EcoRV fragment in the wild-type (wt) genomic DNA, reduced to 7.5 kb in the Af9LZ allele (left). A nonrepetitive 3′ probe consisting of a 1-kb BglII-BamHI fragment (A) detected a 6.5-kb wild-type HindIII band, increased to 11 kb in Af9LZ and then reduced to 10 kb in Af9NO (center). The same 11-kb Af9LZ band hybridized with a probe specific for the inserted neo cassette (right); a single neo insertion was observed. This band was not visible in the Af9NO clones after removal of the neo sequence by expression of Cre recombinase. (C) RT-PCR analysis of RNA from a representative litter of E11.5 Af9NO embryos was used to detect the presence of the Af9 transcript. PCRs were analyzed on agarose gels. PCR carried out with no DNA template (H2O) served as a negative control. A 207-bp band amplified from the Af9 mRNA was generated from all samples, except those from embryos 1, 3, and 4, which were null Af9 mutants. Actin gene-specific RT-PCR, indicating that all cDNA populations analyzed were of comparable quality and concentration, is shown below. Embryo numbers are indicated above the gel. Embryos 1, 3, and 4 were Af9NO−/−, while the remaining embryos were either Af9NO+/− (2, 5, and 7 to 9) or wild type (6 and 10).