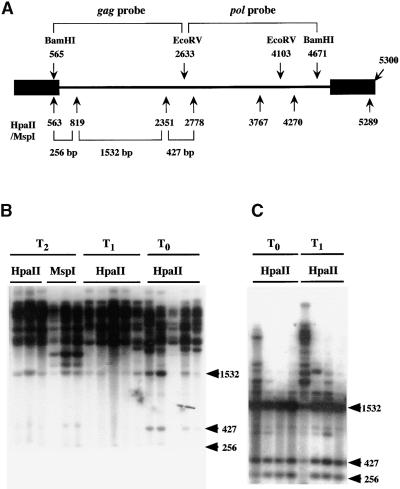
Figure 4.
Analysis of DNA Methylation of Tto1 in T0, T1, and T2 Transgenic Lines.
(A) Restriction maps of Tto1-1 and probes used for the analysis. Black rectangles denote the long terminal repeat.
(B) DNA gel blot analysis of methylation of Tto1 in transgenic lines having high copy numbers. Genomic DNA was digested with HpaII or MspI and analyzed by DNA gel blotting with the Tto1 gag probe. T2 DNA was prepared from the pooled progeny (four regenerated T2 plants from each T1 plant used in Figure 3). T1 DNA was prepared from the pooled progeny (five T1 plants) derived from each T0 line.
(C) DNA gel blot analysis of methylation of Tto1 in transgenic lines having low or medium copy numbers. Genomic DNA was digested with HpaII and analyzed by DNA gel blotting with the Tto1 gag probe. T1 DNA was prepared from the pooled progeny (10 T1 plants) derived from each T0 line. The copy numbers of Tto1 in transgenic lines (from left to right) are six, two, one, and one (data not shown). Only the left-most transgenic line carries transposed copies (four copies).
All of the DNA samples analyzed in (B) and (C) were shown to be equally well digested with the restriction enzyme by reprobing the blot with a single-copy sequence (m105; Pruitt and Meyerowitz, 1986) (data not shown). Lengths and positions of fragments expected from digestion of unmethylated Tto1 are shown at right in (B) and (C) in kilobases.