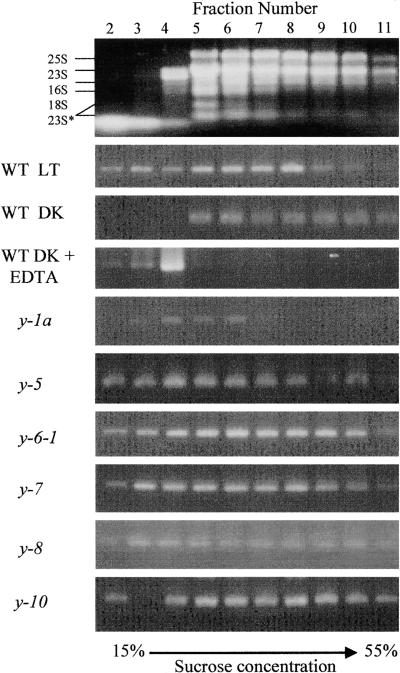
Figure 3.
Association of chlL Transcripts with Polysomes in Light- and Dark-Grown Chlamydomonas Cells.
After sucrose gradient sedimentation of crude polysomal preparations isolated from light- (LT) and dark-grown (DK) wild-type (WT) and y mutant strains of Chlamydomonas, the gradients were divided into 12 fractions. An equal portion of the RNA purified from each fraction was analyzed by semiquantitative RT-PCR to determine the relative abundance of chlL transcripts. After PCR amplification, the reaction products were fractionated by agarose gel electrophoresis and stained with ethidium bromide. The first fraction, which represents the cell extract overlying the gradient, was typically void of RNA after centrifugation, and fraction 12 contained a debris pellet; neither fraction is presented here. At top is the distribution of rRNAs in the various fractions, with the sizes of the rRNAs labeled at left. The effects of EDTA treatment on transcript distribution are also shown. The 25S and 18S are cytoplasmic rRNAs, the 23S and 16S are plastid rRNAs, and the 23S* represents breakdown products of 23S (Rott et al., 1998).