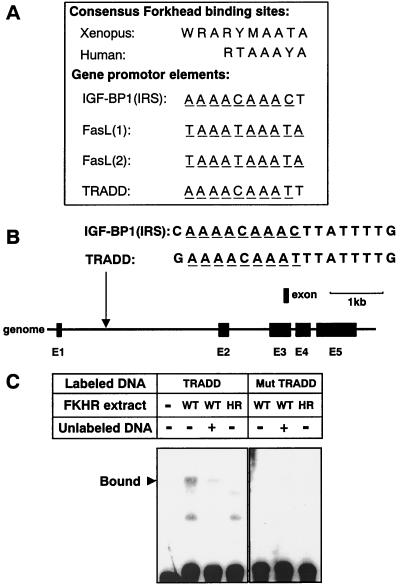
FIG. 6.
Identification of a Forkhead-responsive element in the tradd gene promoter. (A) Comparison of the Xenopus and human consensus Forkhead-responsive element and the Forkhead-responsive element found in the IGF-BP1 (IRS), FasL, or tradd promoter (R = A/G, Y = T/C, W = A/T, M = A/C). The underlined nucleotides matched the predicted Forkhead consensus sequence. (B) Genomic structure of the tradd gene. Black boxes indicate the locations and relative sizes of the five exons. The location of a potential FKHR-binding sequence is indicated in intron 1 and is compared with the consensus FKHR-binding sequence of IGF-BP1 (IRS). (C) Nuclear extracts of HT1080 cells that had been transfected with vector pcDNA3 encoding WT-FKHR (WT) or H215R-FKHR (HR) were incubated with biotin-labeled double-stranded oligonucleotides 5′-biotin-TGAAAACAAATTTATTTTGGGTAC-3′ (TRADD) and 5′-biotin-TGAAACCAAAGTCATTTTGGGTAC-3′ (Mut TRADD) (Forkhead binding sites are underlined). In some experiments, reactions were performed in the presence (+) or absence (−) of a 100-fold excess of unlabeled tradd oligonucleotide (5′-TGAAAACAAATTTATTTTGGGTAC-3′). DNA-protein complexes were fractionated by polyacrylamide gel electrophoresis and visualized by horseradish peroxidase-conjugated streptavidin. The specific DNA-protein complex is indicated on the left.