Figure 2.
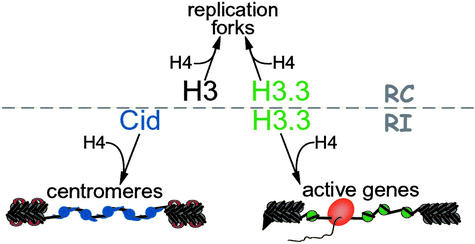
The three versions of histone H3 determine the mode of nucleosome assembly. The deposition of H3 is strictly replication-coupled (RC), and H3 is recruited to replication forks for chromatin doubling. Deposition of Cid (blue) is exclusively replication-independent (RI), and normally occurs only at centromeres. H3.3 (green) undergoes RC deposition, and RI deposition at active loci. Open chromatin at centromeres and at active genes may promote histone replacement. Transcriptional activity, chromatin remodeling factors, and RNA polymerases (orange) will unfold the chromatin fiber and disrupt nucleosome (gray) structure (open chromatin). Transcriptionally inactive regions are not subjected to these forces and remain in a closed configuration. Flanking heterochromatin and H3-containing blocks within the centromeric domain presents the H3K9me epitope, thereby binding HP1 (red) and resulting in a compacted, closed chromatin structure. Cid-containing nucleosomes cannot be methylated in this way, and thus remain comparatively open. The specialized N-terminal tail of Cid may alter the linker DNA between nucleosomes, also contributing to the open chromatin configuration. RI deposition of H3.3 is limited to the open chromatin at active genes and RI deposition of Cid is limited to the open chromatin in centromeric domains.