Figure 4.
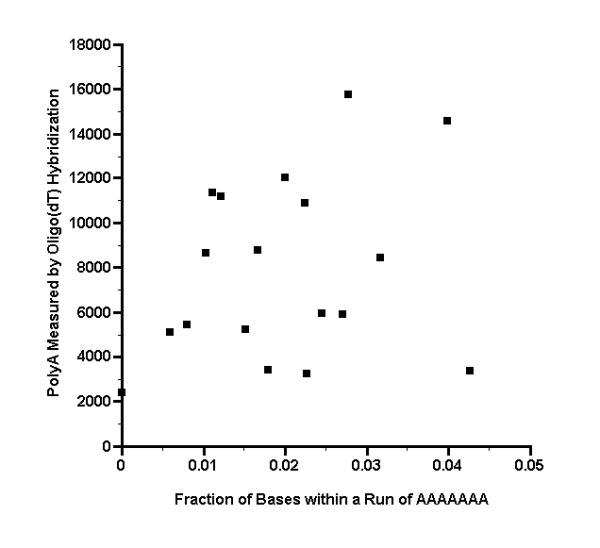
Hybridization intensity from oligo(dT) probe, as a function of the fraction of bases that are within a sequence run of AAAAAAA Oligo(dT) was end labeled with T4 kinase, then hybridized to a microarray. The complete sequences of 18 clone inserts were then determined, corresponding to microarray spots that had a signal of >24000 phosphorimager units with a muscle cDNA probe (see Additional File 1). Each sequence was examined on both strands to determine the number of bases that lie within a run of seven or more As (AAAAAAA), which is a run length that might form a stable hybrid with oligo(dT). The total number of such bases was then divided by twice the sequence length to obtain the fraction of bases that are within a sequence run of AAAAAAA. The fractions are plotted here versus the hybridization intensity from the oligo(dT) probe.
